Chlorine »
PDB 7w4o-7wl9 »
7wf6 »
Chlorine in PDB 7wf6: Crystal Structure of SNX13 Rgs Domain
Protein crystallography data
The structure of Crystal Structure of SNX13 Rgs Domain, PDB code: 7wf6
was solved by
J.Xu,
J.Zhu,
J.Liu,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 18.00 / 3.25 |
| Space group | P 43 3 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 165.328, 165.328, 165.328, 90, 90, 90 |
| R / Rfree (%) | 21.9 / 22.4 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of SNX13 Rgs Domain
(pdb code 7wf6). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 4 binding sites of Chlorine where determined in the Crystal Structure of SNX13 Rgs Domain, PDB code: 7wf6:
Jump to Chlorine binding site number: 1; 2; 3; 4;
In total 4 binding sites of Chlorine where determined in the Crystal Structure of SNX13 Rgs Domain, PDB code: 7wf6:
Jump to Chlorine binding site number: 1; 2; 3; 4;
Chlorine binding site 1 out of 4 in 7wf6
Go back to
Chlorine binding site 1 out
of 4 in the Crystal Structure of SNX13 Rgs Domain

Mono view
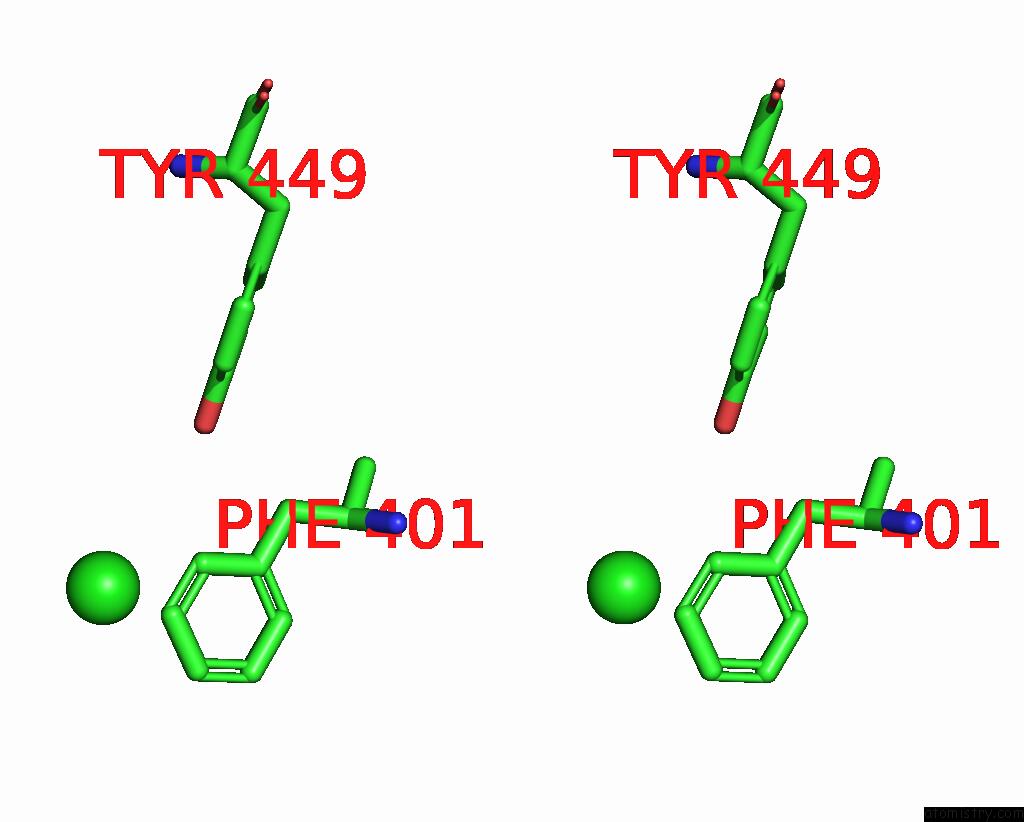
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of SNX13 Rgs Domain within 5.0Å range:
|
Chlorine binding site 2 out of 4 in 7wf6
Go back to
Chlorine binding site 2 out
of 4 in the Crystal Structure of SNX13 Rgs Domain
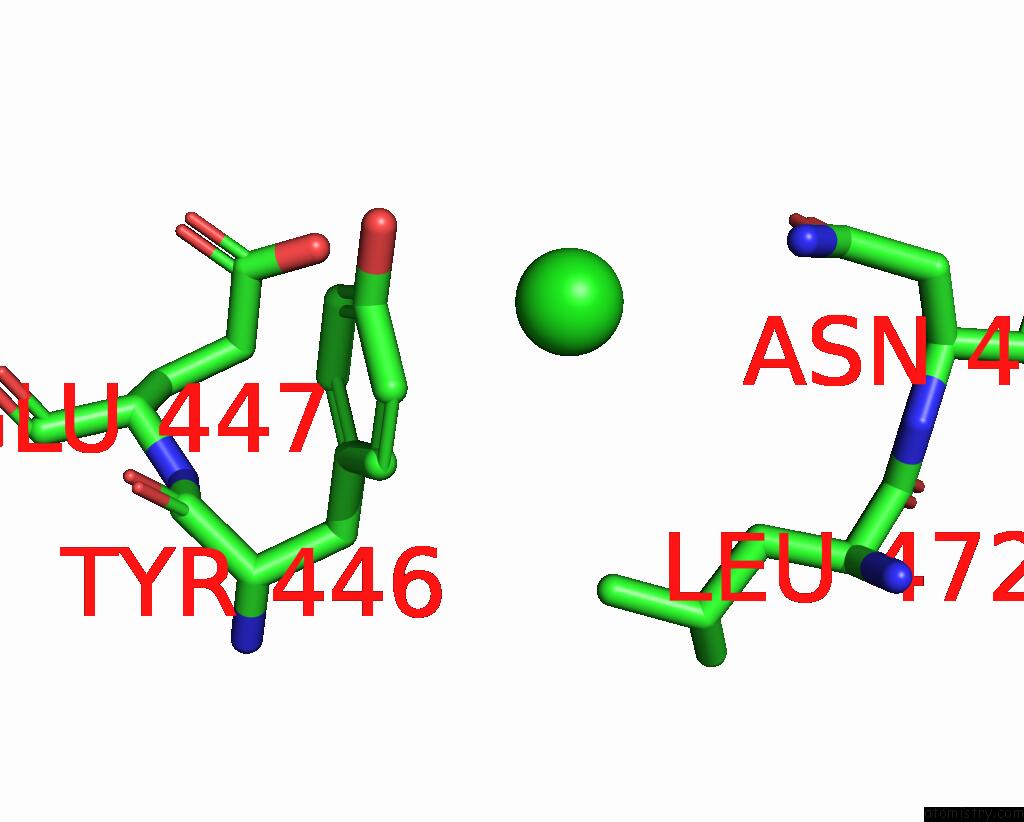
Mono view
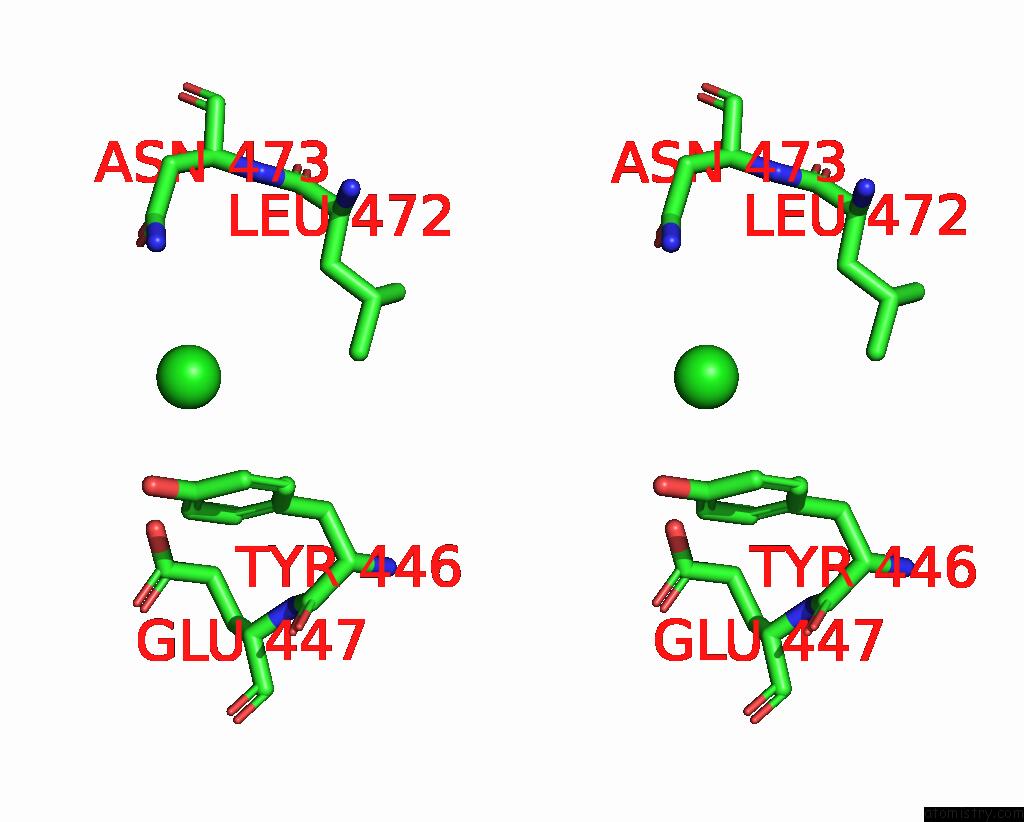
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of SNX13 Rgs Domain within 5.0Å range:
|
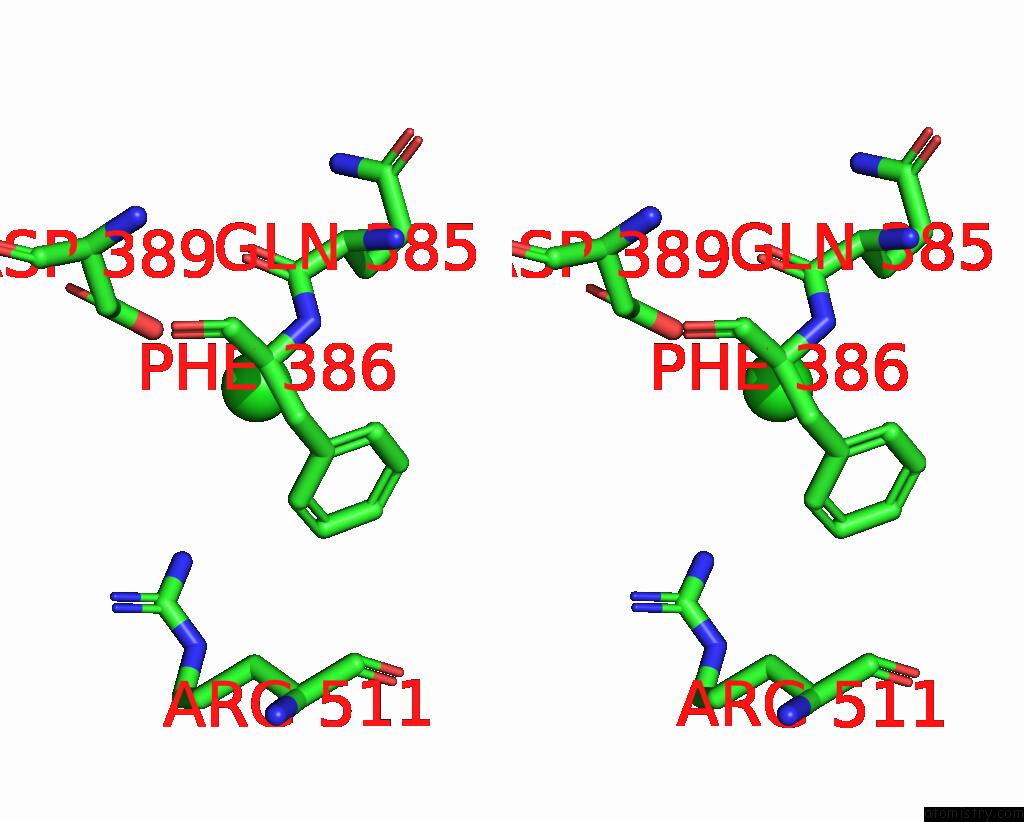
Chlorine binding site 3 out of 4 in 7wf6
Go back to
Chlorine binding site 3 out
of 4 in the Crystal Structure of SNX13 Rgs Domain
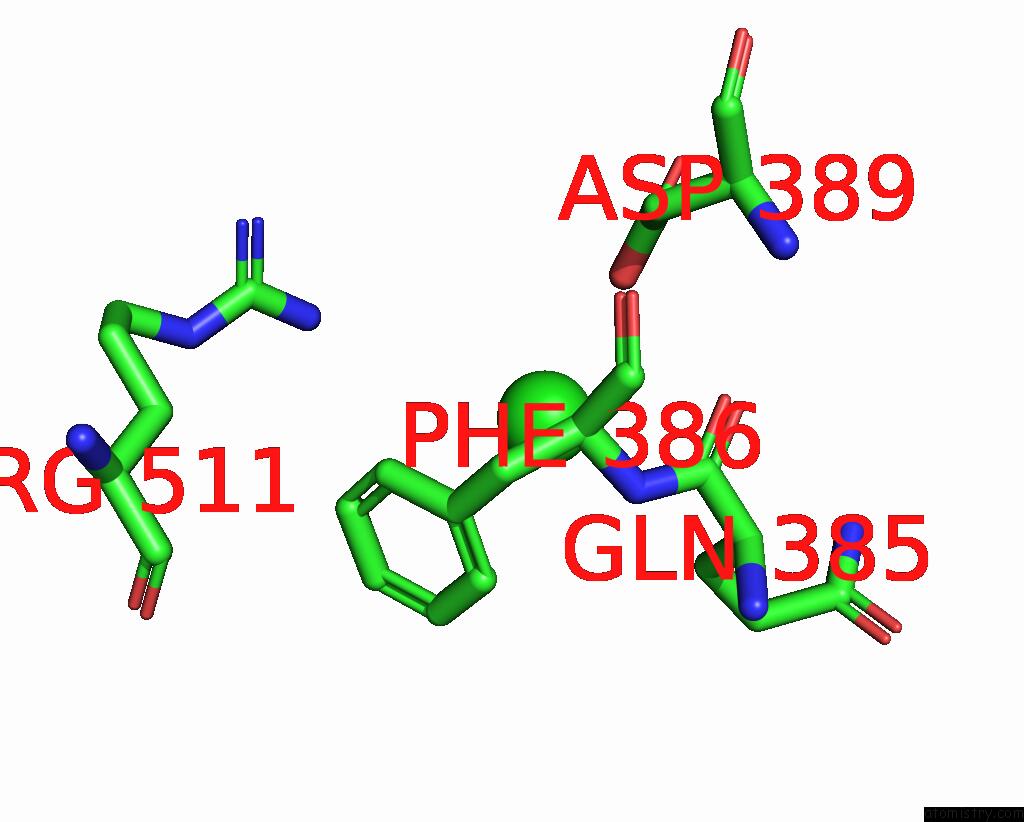
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Crystal Structure of SNX13 Rgs Domain within 5.0Å range:
|
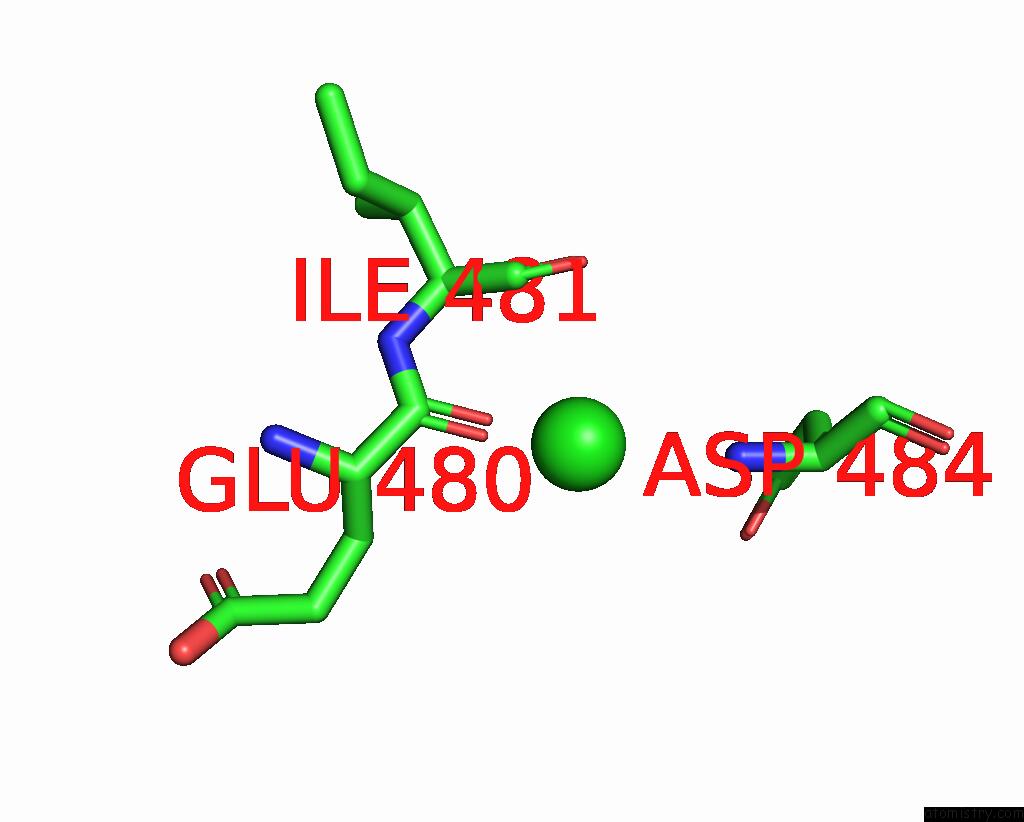
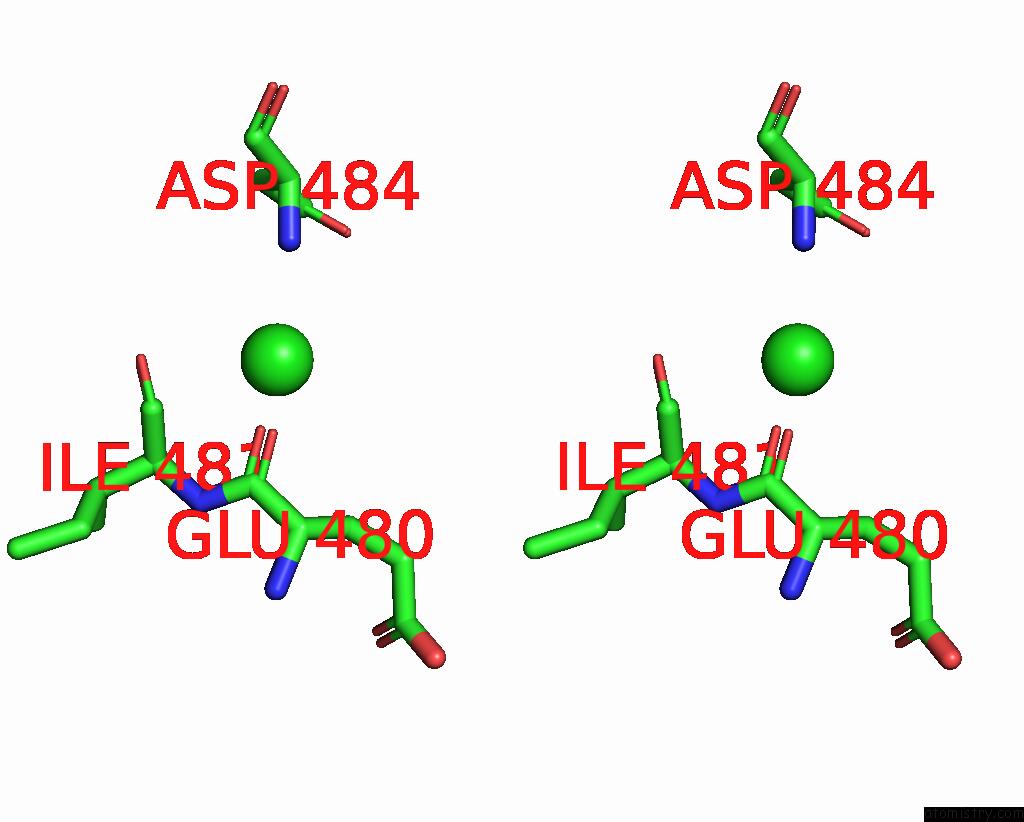
Chlorine binding site 4 out of 4 in 7wf6
Go back to
Chlorine binding site 4 out
of 4 in the Crystal Structure of SNX13 Rgs Domain
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Crystal Structure of SNX13 Rgs Domain within 5.0Å range:
|
Reference:
Y.Zhang,
R.Chen,
Y.Dong,
J.Zhu,
K.Su,
J.Liu,
J.Xu.
Structural Studies Reveal Unique Non-Canonical Regulators of G Protein Signaling Homology (Rh) Domains in Sorting Nexins. J.Mol.Biol. V. 434 67823 2022.
ISSN: ESSN 1089-8638
PubMed: 36103920
DOI: 10.1016/J.JMB.2022.167823
Page generated: Sun Jul 13 08:17:21 2025
ISSN: ESSN 1089-8638
PubMed: 36103920
DOI: 10.1016/J.JMB.2022.167823
Last articles
Mo in 3ETRMo in 3FAH
Mo in 3BDJ
Mo in 3EGW
Mo in 3DMR
Mo in 3B9J
Mo in 3AX9
Mo in 3AX7
Mo in 3AMZ
Mo in 3AM9