Chlorine »
PDB 7xqx-7ycb »
7y1g »
Chlorine in PDB 7y1g: Crystal Structure of Human Prkaca Complexed with DS01080522
Enzymatic activity of Crystal Structure of Human Prkaca Complexed with DS01080522
All present enzymatic activity of Crystal Structure of Human Prkaca Complexed with DS01080522:
2.7.11.11;
2.7.11.11;
Protein crystallography data
The structure of Crystal Structure of Human Prkaca Complexed with DS01080522, PDB code: 7y1g
was solved by
M.Suzuki,
O.Ubukata,
A.Toyoda,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 25.00 / 2.30 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 92.221, 87.63, 113.476, 90, 106.72, 90 |
| R / Rfree (%) | 18.6 / 23.7 |
Other elements in 7y1g:
The structure of Crystal Structure of Human Prkaca Complexed with DS01080522 also contains other interesting chemical elements:
| Zinc | (Zn) | 1 atom |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of Human Prkaca Complexed with DS01080522
(pdb code 7y1g). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 4 binding sites of Chlorine where determined in the Crystal Structure of Human Prkaca Complexed with DS01080522, PDB code: 7y1g:
Jump to Chlorine binding site number: 1; 2; 3; 4;
In total 4 binding sites of Chlorine where determined in the Crystal Structure of Human Prkaca Complexed with DS01080522, PDB code: 7y1g:
Jump to Chlorine binding site number: 1; 2; 3; 4;
Chlorine binding site 1 out of 4 in 7y1g
Go back to
Chlorine binding site 1 out
of 4 in the Crystal Structure of Human Prkaca Complexed with DS01080522
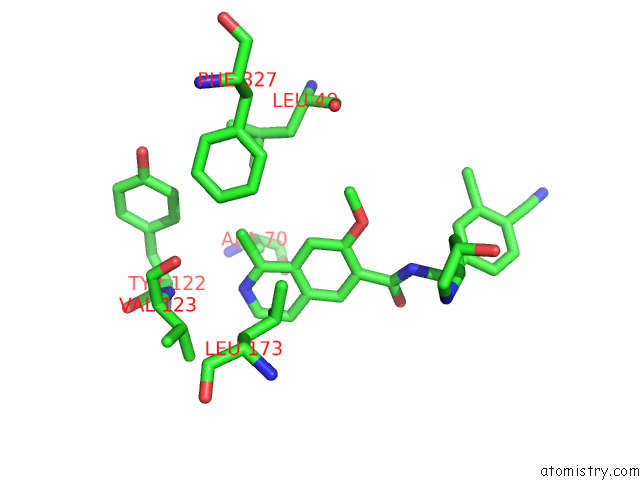
Mono view
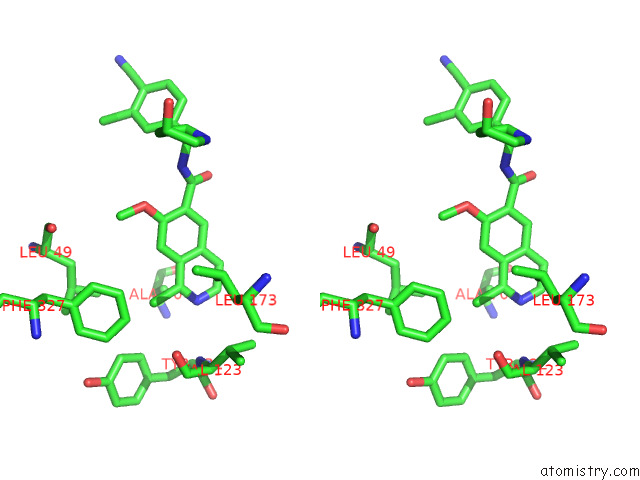
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Human Prkaca Complexed with DS01080522 within 5.0Å range:
|
Chlorine binding site 2 out of 4 in 7y1g
Go back to
Chlorine binding site 2 out
of 4 in the Crystal Structure of Human Prkaca Complexed with DS01080522
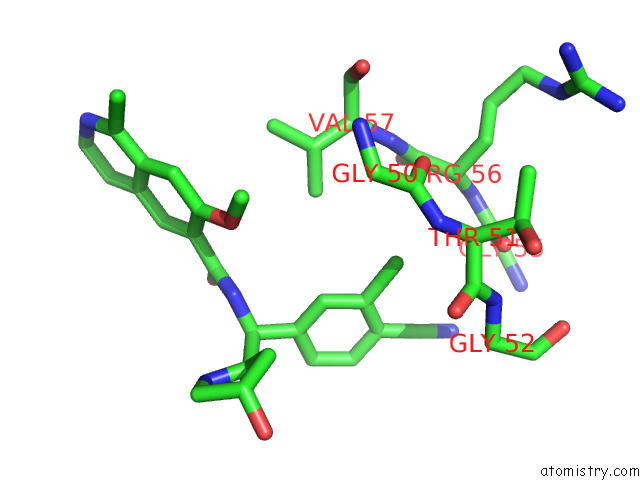
Mono view
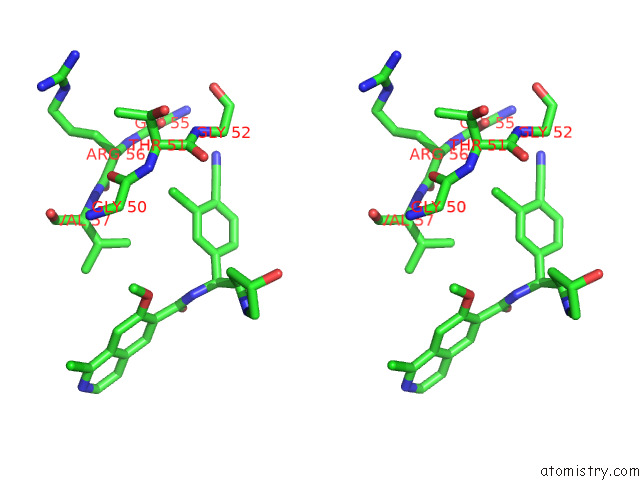
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of Human Prkaca Complexed with DS01080522 within 5.0Å range:
|
Chlorine binding site 3 out of 4 in 7y1g
Go back to
Chlorine binding site 3 out
of 4 in the Crystal Structure of Human Prkaca Complexed with DS01080522
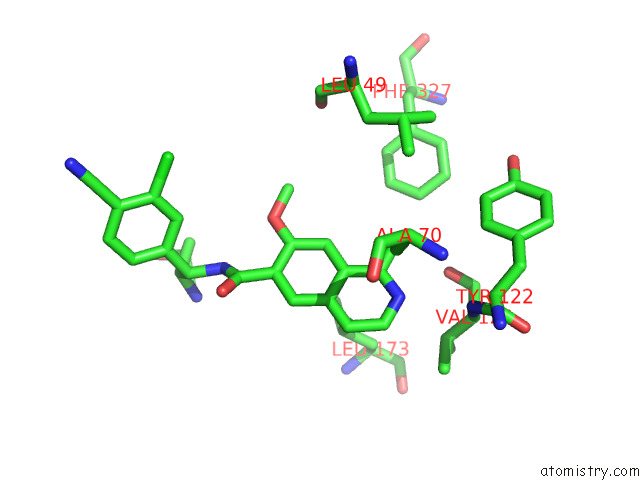
Mono view
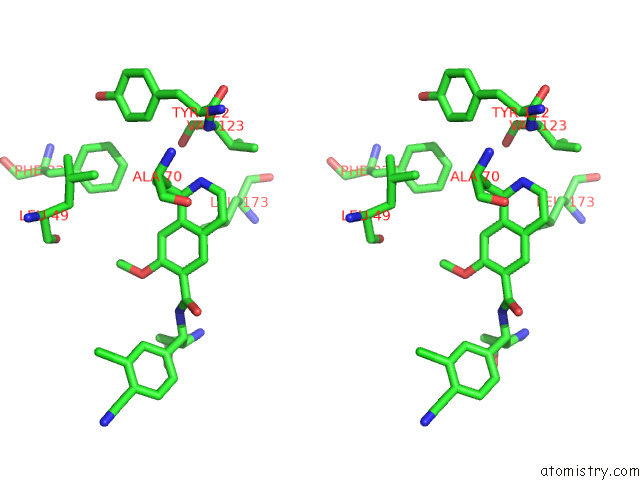
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Crystal Structure of Human Prkaca Complexed with DS01080522 within 5.0Å range:
|
Chlorine binding site 4 out of 4 in 7y1g
Go back to
Chlorine binding site 4 out
of 4 in the Crystal Structure of Human Prkaca Complexed with DS01080522
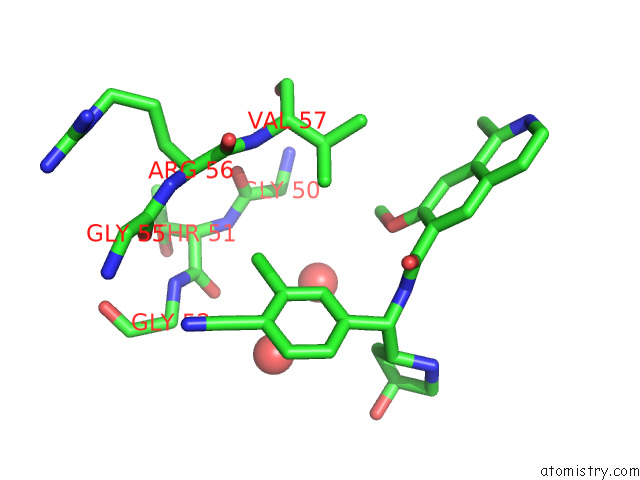
Mono view
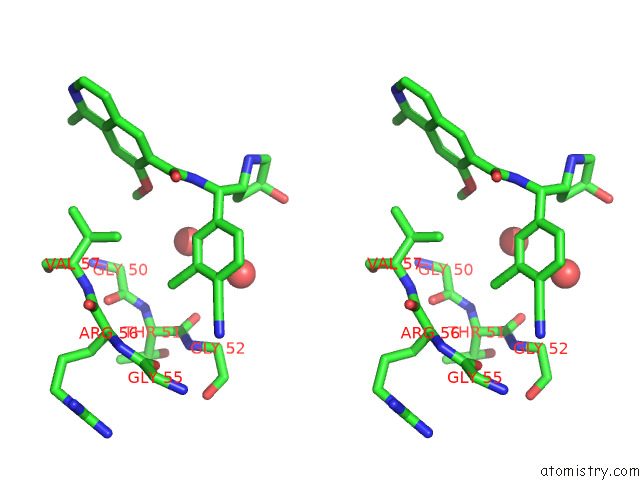
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Crystal Structure of Human Prkaca Complexed with DS01080522 within 5.0Å range:
|
Reference:
A.Toyota,
M.Goto,
M.Miyamoto,
Y.Nagashima,
S.Iwasaki,
T.Komatsu,
T.Momose,
K.Yoshida,
T.Tsukada,
T.Matsufuji,
A.Ohno,
M.Suzuki,
O.Ubukata,
Y.Kaneta.
Novel Protein Kinase Camp-Activated Catalytic Subunit Alpha (Prkaca) Inhibitor Shows Anti-Tumor Activity in A Fibrolamellar Hepatocellular Carcinoma Model. Biochem.Biophys.Res.Commun. V. 621 157 2022.
ISSN: ESSN 1090-2104
PubMed: 35839742
DOI: 10.1016/J.BBRC.2022.07.008
Page generated: Sun Jul 13 08:31:16 2025
ISSN: ESSN 1090-2104
PubMed: 35839742
DOI: 10.1016/J.BBRC.2022.07.008
Last articles
Mn in 3NIOMn in 3NWK
Mn in 3O1R
Mn in 3NVT
Mn in 3O1P
Mn in 3O1O
Mn in 3O1M
Mn in 3NV8
Mn in 3NUE
Mn in 3NQB