Chlorine »
PDB 7z21-7zga »
7z53 »
Chlorine in PDB 7z53: Structure of Native Leukocyte Myeloperoxidase in Complex with A Truncated Version (Spin Truncated) of the Staphyloccal Peroxidase Inhibitor Spin From Staphylococcus Aureus
Protein crystallography data
The structure of Structure of Native Leukocyte Myeloperoxidase in Complex with A Truncated Version (Spin Truncated) of the Staphyloccal Peroxidase Inhibitor Spin From Staphylococcus Aureus, PDB code: 7z53
was solved by
V.Pfanzagl,
J.A.Brito,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 76.19 / 2.28 |
| Space group | P 21 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 257.022, 157.196, 166.56, 90, 90, 90 |
| R / Rfree (%) | 17.4 / 22.9 |
Other elements in 7z53:
The structure of Structure of Native Leukocyte Myeloperoxidase in Complex with A Truncated Version (Spin Truncated) of the Staphyloccal Peroxidase Inhibitor Spin From Staphylococcus Aureus also contains other interesting chemical elements:
| Iron | (Fe) | 8 atoms |
| Calcium | (Ca) | 8 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Structure of Native Leukocyte Myeloperoxidase in Complex with A Truncated Version (Spin Truncated) of the Staphyloccal Peroxidase Inhibitor Spin From Staphylococcus Aureus
(pdb code 7z53). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 8 binding sites of Chlorine where determined in the Structure of Native Leukocyte Myeloperoxidase in Complex with A Truncated Version (Spin Truncated) of the Staphyloccal Peroxidase Inhibitor Spin From Staphylococcus Aureus, PDB code: 7z53:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
In total 8 binding sites of Chlorine where determined in the Structure of Native Leukocyte Myeloperoxidase in Complex with A Truncated Version (Spin Truncated) of the Staphyloccal Peroxidase Inhibitor Spin From Staphylococcus Aureus, PDB code: 7z53:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
Chlorine binding site 1 out of 8 in 7z53
Go back to
Chlorine binding site 1 out
of 8 in the Structure of Native Leukocyte Myeloperoxidase in Complex with A Truncated Version (Spin Truncated) of the Staphyloccal Peroxidase Inhibitor Spin From Staphylococcus Aureus
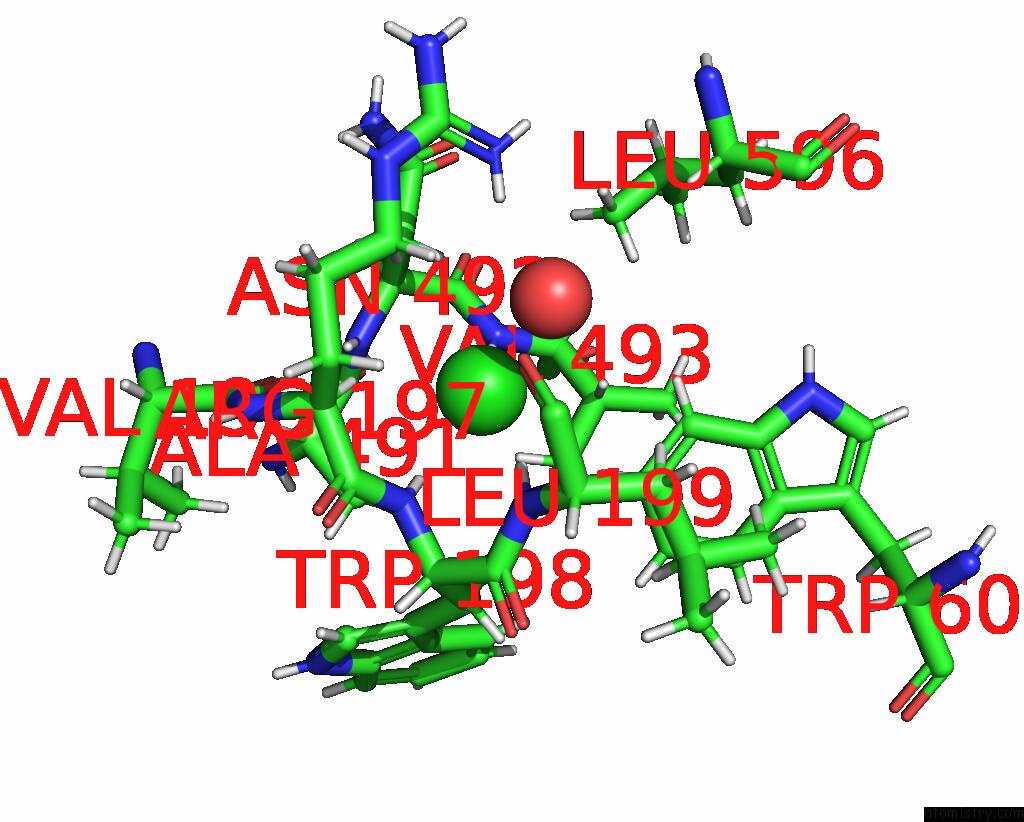
Mono view
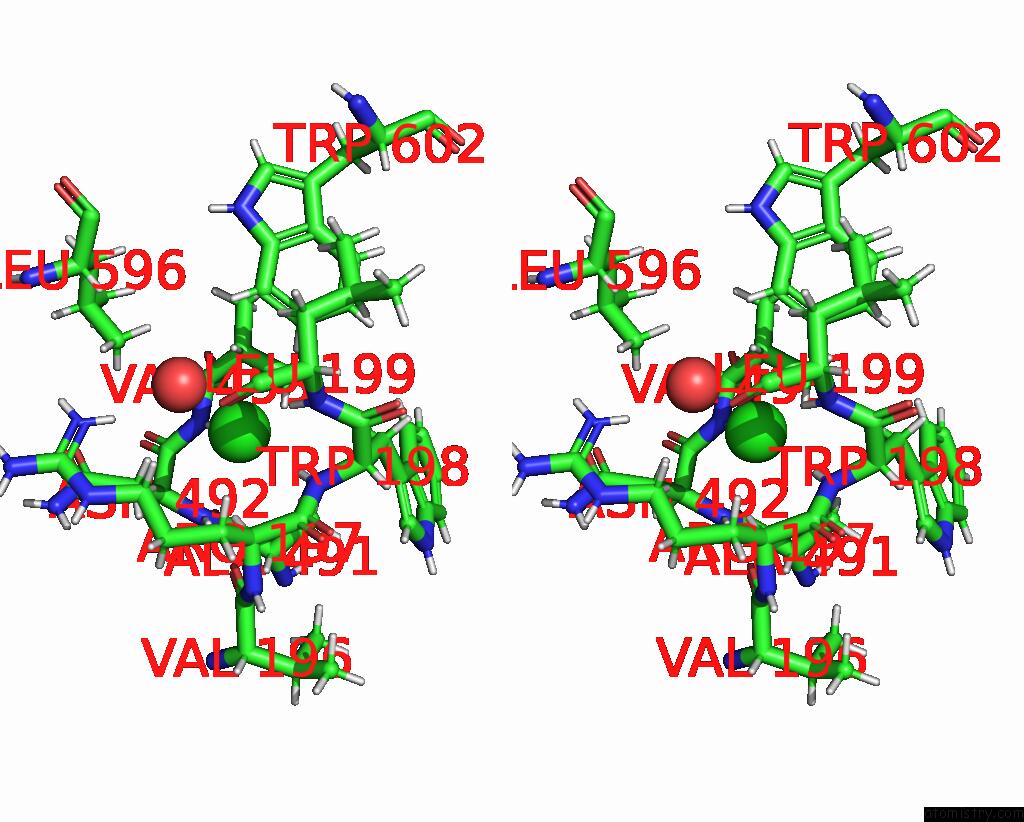
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Structure of Native Leukocyte Myeloperoxidase in Complex with A Truncated Version (Spin Truncated) of the Staphyloccal Peroxidase Inhibitor Spin From Staphylococcus Aureus within 5.0Å range:
|
Chlorine binding site 2 out of 8 in 7z53
Go back to
Chlorine binding site 2 out
of 8 in the Structure of Native Leukocyte Myeloperoxidase in Complex with A Truncated Version (Spin Truncated) of the Staphyloccal Peroxidase Inhibitor Spin From Staphylococcus Aureus
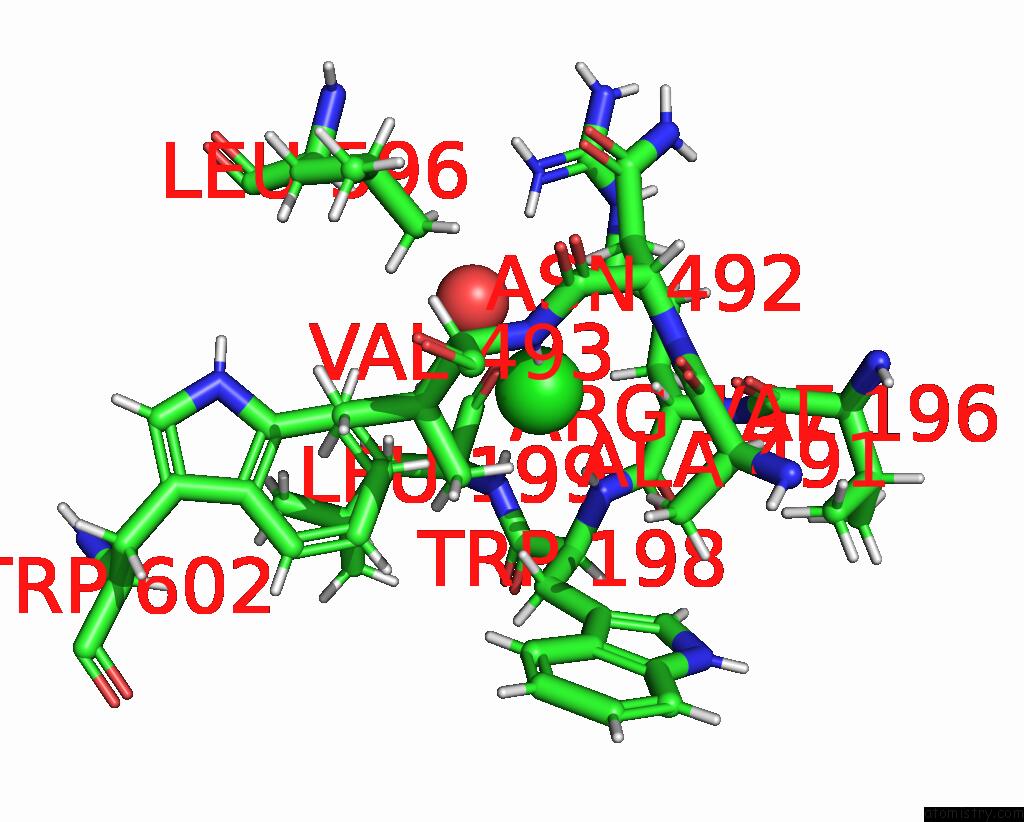
Mono view
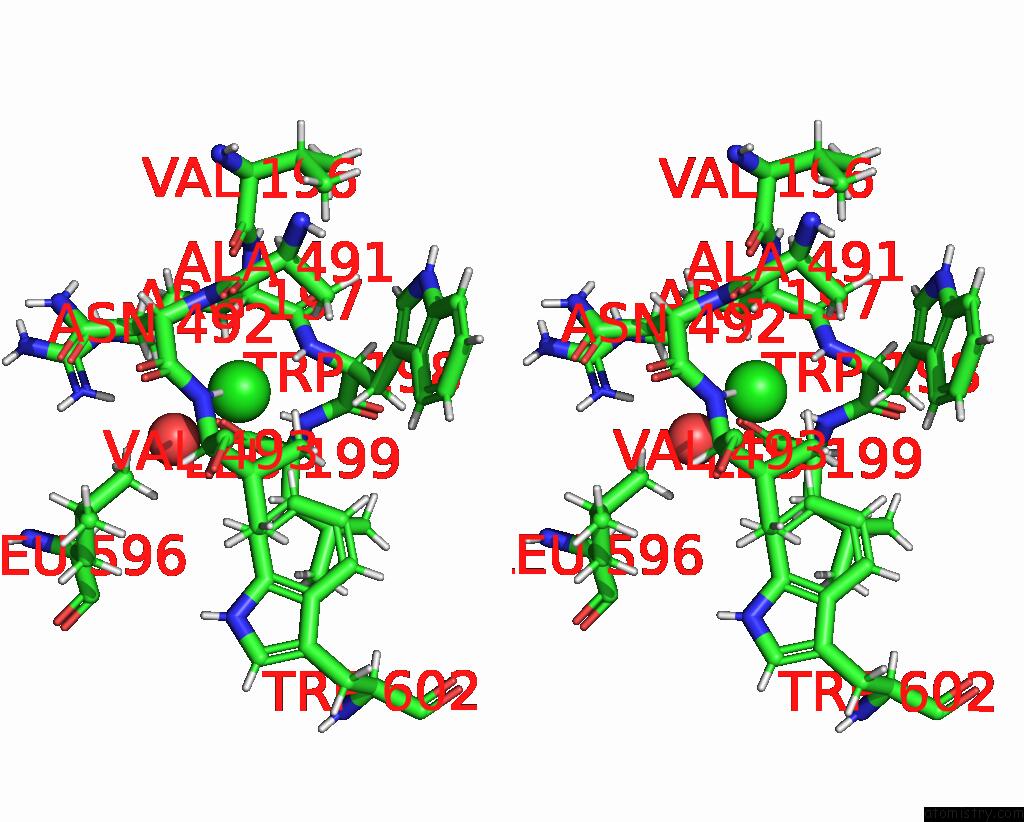
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Structure of Native Leukocyte Myeloperoxidase in Complex with A Truncated Version (Spin Truncated) of the Staphyloccal Peroxidase Inhibitor Spin From Staphylococcus Aureus within 5.0Å range:
|
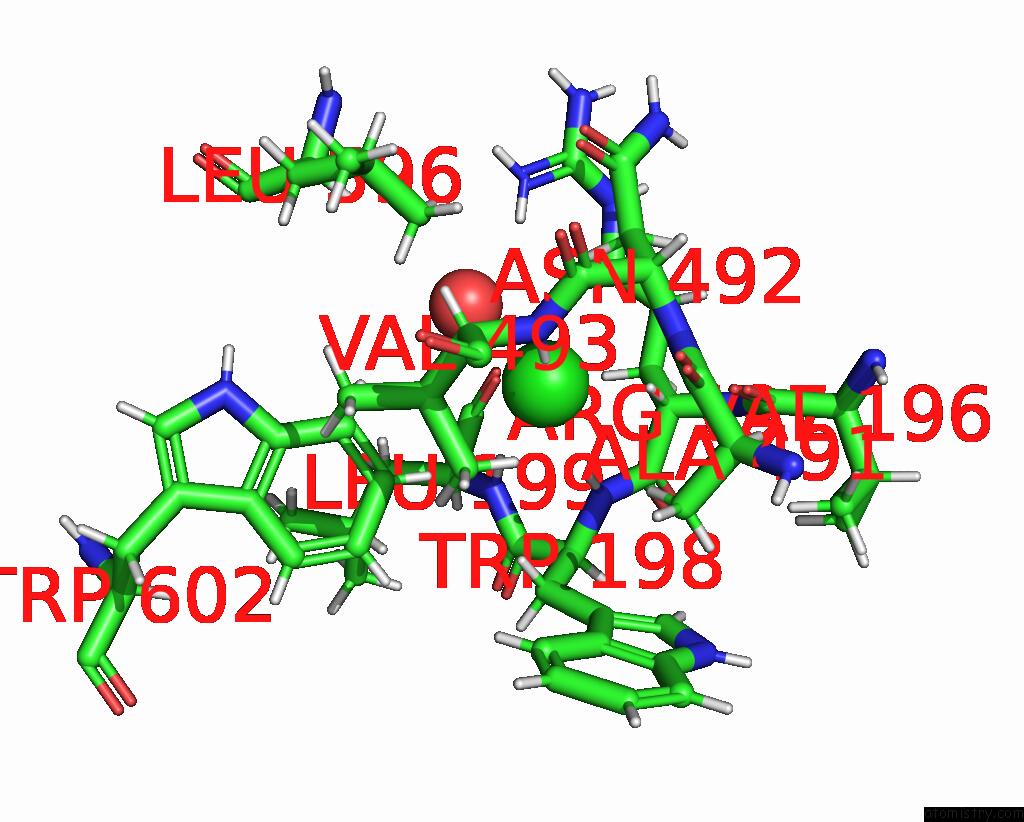
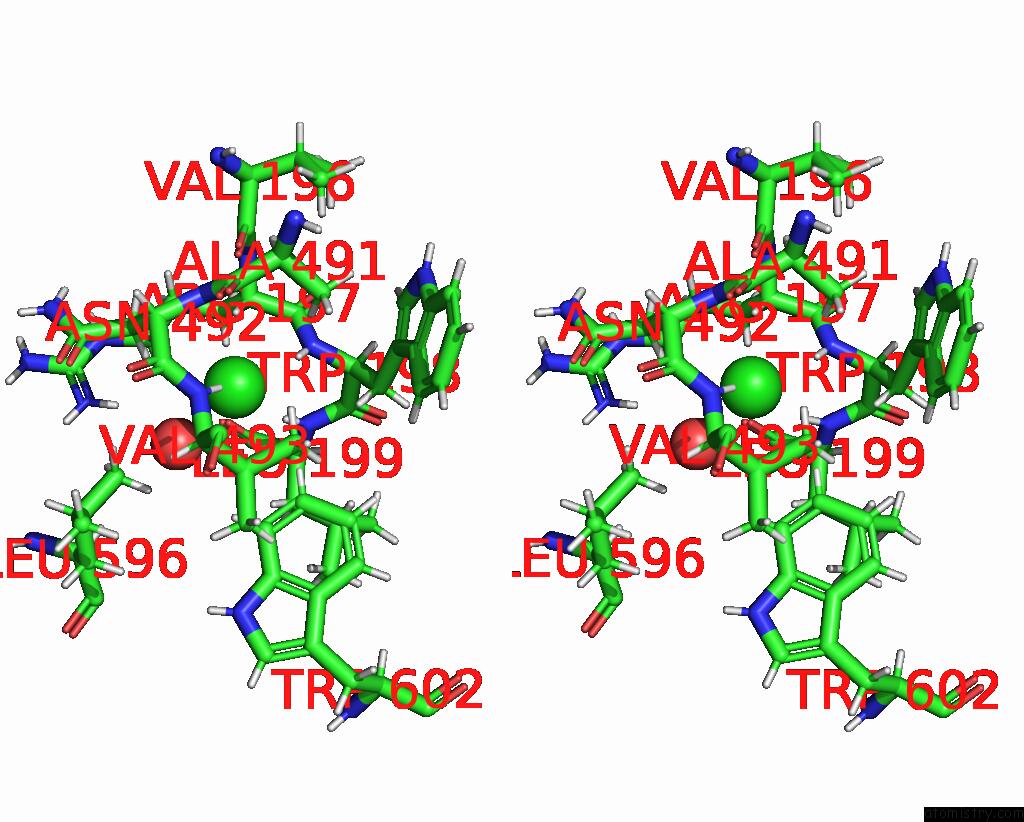
Chlorine binding site 3 out of 8 in 7z53
Go back to
Chlorine binding site 3 out
of 8 in the Structure of Native Leukocyte Myeloperoxidase in Complex with A Truncated Version (Spin Truncated) of the Staphyloccal Peroxidase Inhibitor Spin From Staphylococcus Aureus
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Structure of Native Leukocyte Myeloperoxidase in Complex with A Truncated Version (Spin Truncated) of the Staphyloccal Peroxidase Inhibitor Spin From Staphylococcus Aureus within 5.0Å range:
|
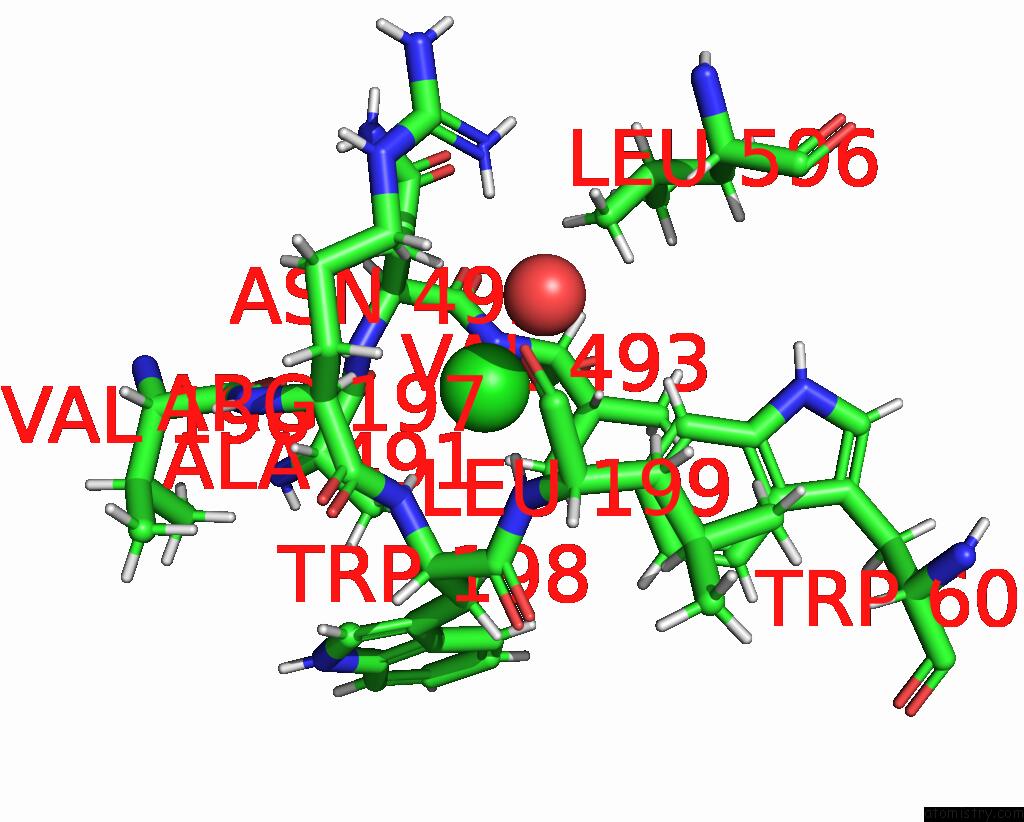
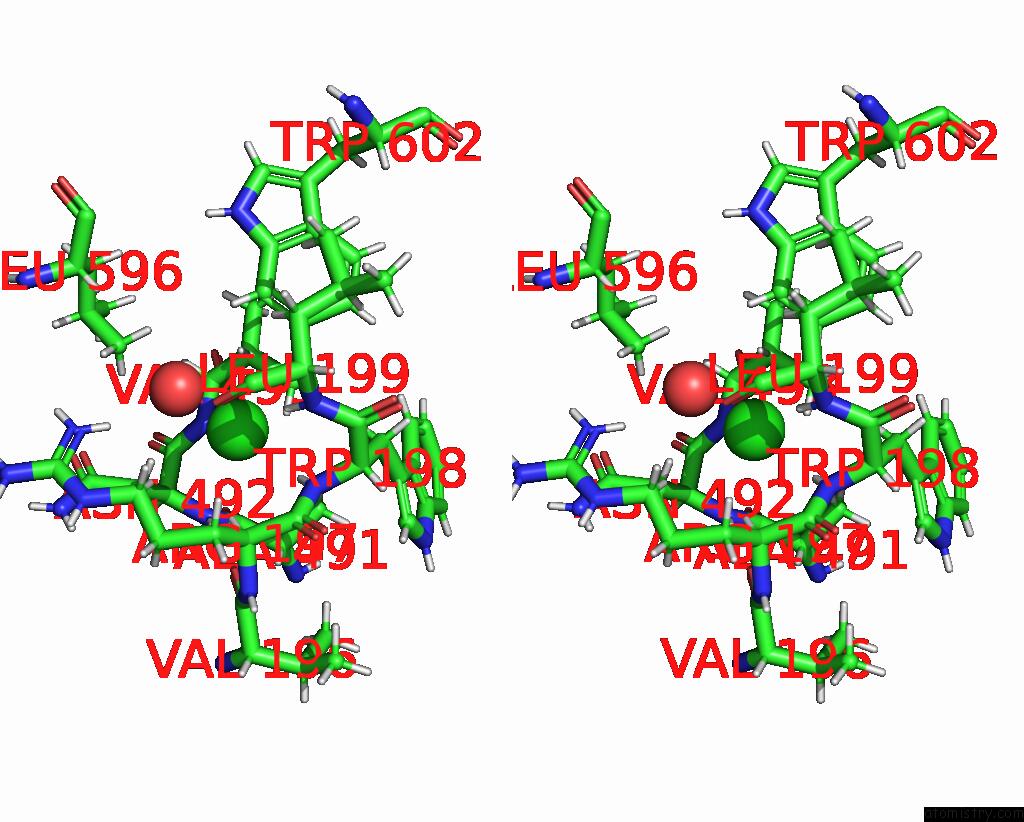
Chlorine binding site 4 out of 8 in 7z53
Go back to
Chlorine binding site 4 out
of 8 in the Structure of Native Leukocyte Myeloperoxidase in Complex with A Truncated Version (Spin Truncated) of the Staphyloccal Peroxidase Inhibitor Spin From Staphylococcus Aureus
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Structure of Native Leukocyte Myeloperoxidase in Complex with A Truncated Version (Spin Truncated) of the Staphyloccal Peroxidase Inhibitor Spin From Staphylococcus Aureus within 5.0Å range:
|
Chlorine binding site 5 out of 8 in 7z53
Go back to
Chlorine binding site 5 out
of 8 in the Structure of Native Leukocyte Myeloperoxidase in Complex with A Truncated Version (Spin Truncated) of the Staphyloccal Peroxidase Inhibitor Spin From Staphylococcus Aureus
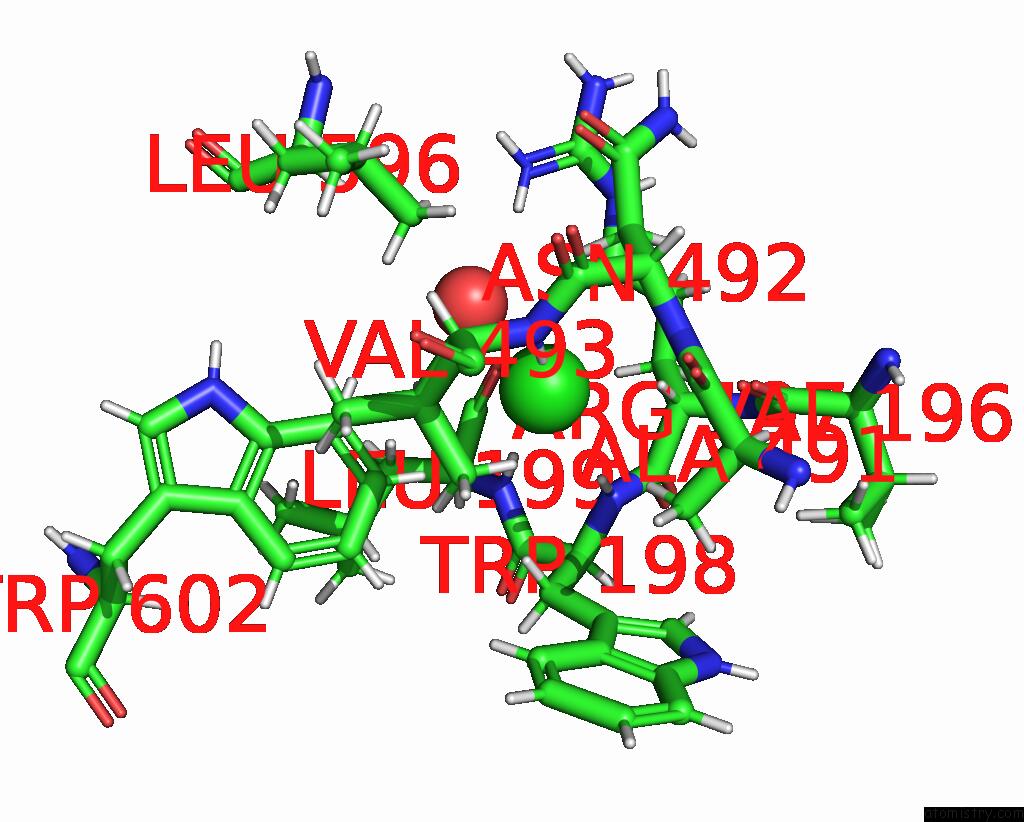
Mono view
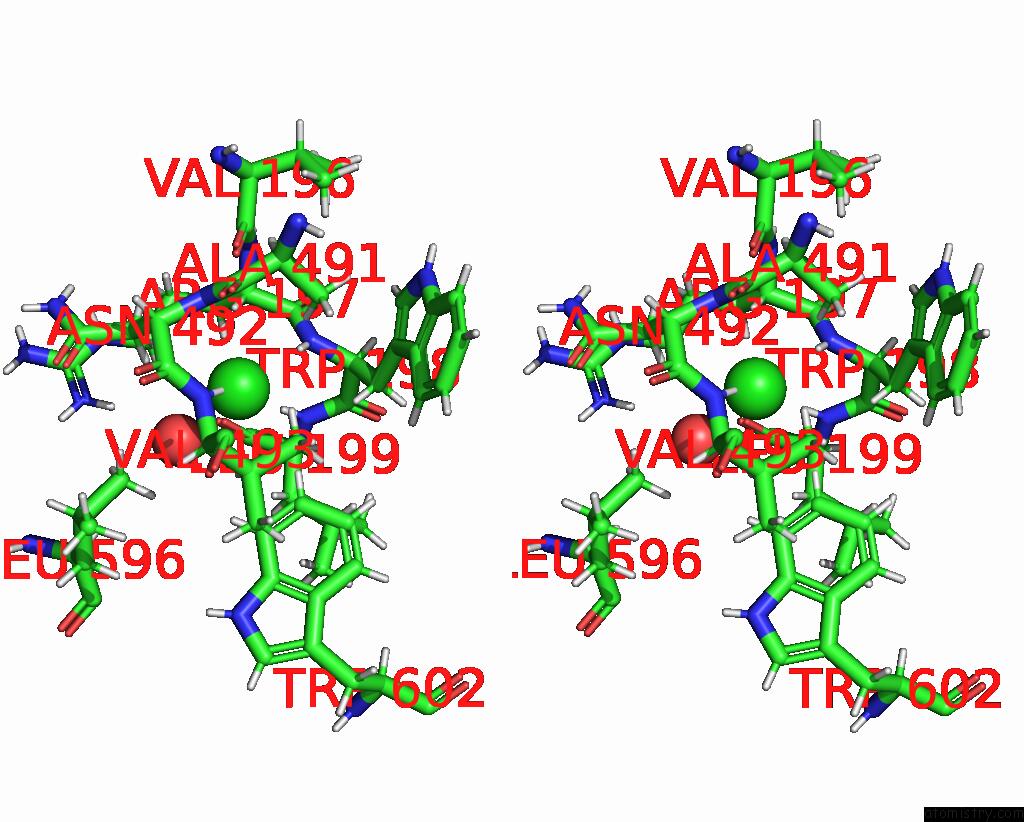
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Structure of Native Leukocyte Myeloperoxidase in Complex with A Truncated Version (Spin Truncated) of the Staphyloccal Peroxidase Inhibitor Spin From Staphylococcus Aureus within 5.0Å range:
|
Chlorine binding site 6 out of 8 in 7z53
Go back to
Chlorine binding site 6 out
of 8 in the Structure of Native Leukocyte Myeloperoxidase in Complex with A Truncated Version (Spin Truncated) of the Staphyloccal Peroxidase Inhibitor Spin From Staphylococcus Aureus
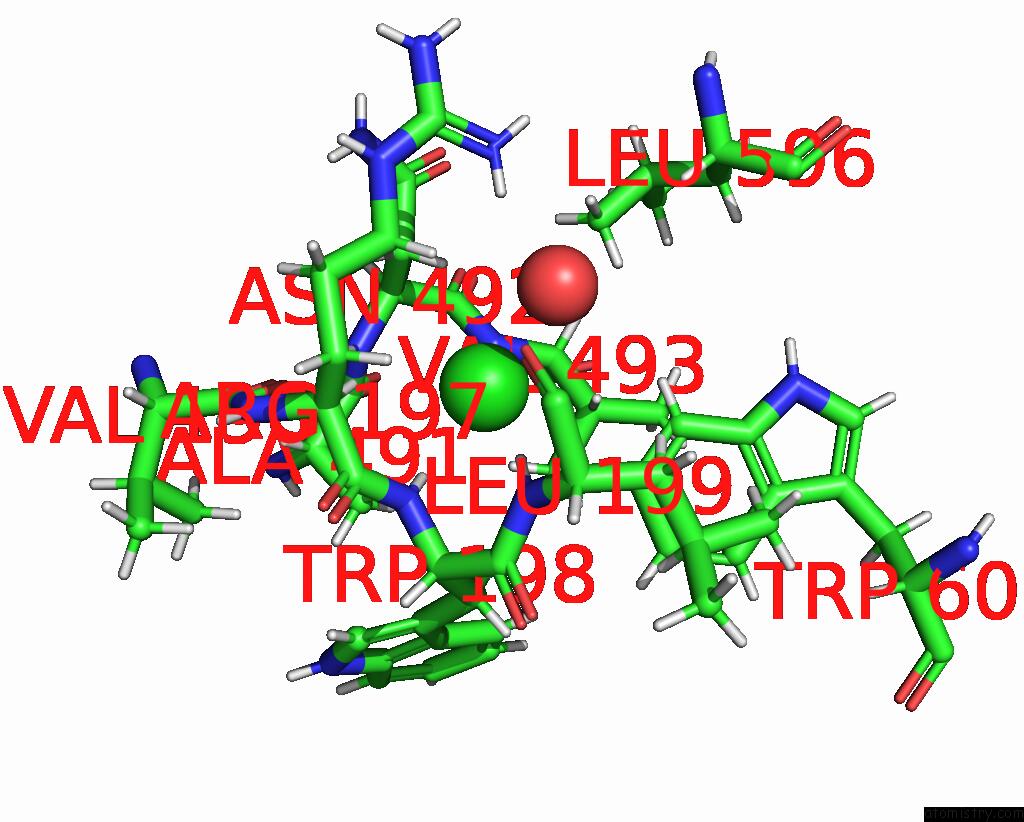
Mono view
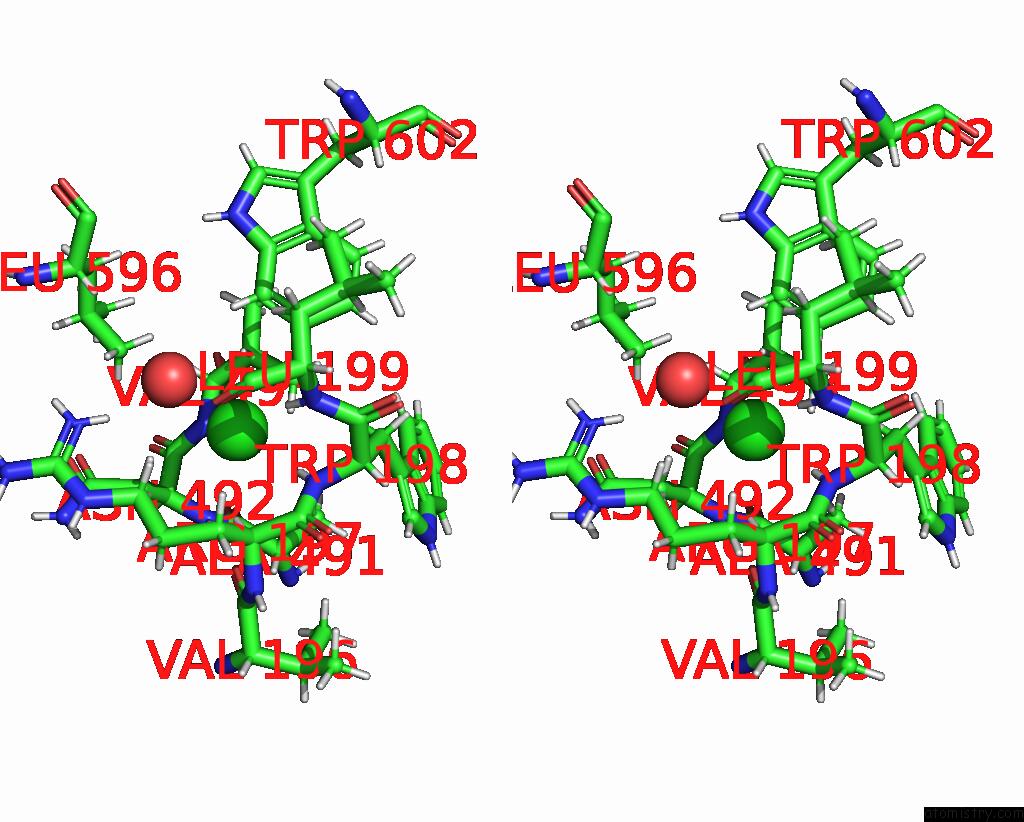
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of Structure of Native Leukocyte Myeloperoxidase in Complex with A Truncated Version (Spin Truncated) of the Staphyloccal Peroxidase Inhibitor Spin From Staphylococcus Aureus within 5.0Å range:
|
Chlorine binding site 7 out of 8 in 7z53
Go back to
Chlorine binding site 7 out
of 8 in the Structure of Native Leukocyte Myeloperoxidase in Complex with A Truncated Version (Spin Truncated) of the Staphyloccal Peroxidase Inhibitor Spin From Staphylococcus Aureus
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 7 of Structure of Native Leukocyte Myeloperoxidase in Complex with A Truncated Version (Spin Truncated) of the Staphyloccal Peroxidase Inhibitor Spin From Staphylococcus Aureus within 5.0Å range:
|
Chlorine binding site 8 out of 8 in 7z53
Go back to
Chlorine binding site 8 out
of 8 in the Structure of Native Leukocyte Myeloperoxidase in Complex with A Truncated Version (Spin Truncated) of the Staphyloccal Peroxidase Inhibitor Spin From Staphylococcus Aureus
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 8 of Structure of Native Leukocyte Myeloperoxidase in Complex with A Truncated Version (Spin Truncated) of the Staphyloccal Peroxidase Inhibitor Spin From Staphylococcus Aureus within 5.0Å range:
|
Reference:
U.Leitgeb,
P.G.Furtmuller,
S.Hofbauer,
J.A.Brito,
C.Obinger,
V.Pfanzagl.
The Staphylococcal Inhibitory Protein Spin Binds to Human Myeloperoxidase with Picomolar Affinity But Only Dampens Halide Oxidation. J.Biol.Chem. V. 298 02514 2022.
ISSN: ESSN 1083-351X
PubMed: 36150500
DOI: 10.1016/J.JBC.2022.102514
Page generated: Sun Jul 13 08:41:48 2025
ISSN: ESSN 1083-351X
PubMed: 36150500
DOI: 10.1016/J.JBC.2022.102514
Last articles
Mg in 2OQYMg in 2OUP
Mg in 2OUN
Mg in 2OU7
Mg in 2OTG
Mg in 2OSB
Mg in 2OS8
Mg in 2ORI
Mg in 2ORW
Mg in 2ONP