Chlorine »
PDB 8cl5-8cur »
8crm »
Chlorine in PDB 8crm: Crystal Structure of N-Terminal Sars-Cov-2 NSP1 in Complex with Fragment Hit 11C6 Refined Against Anomalous Diffraction Data
Protein crystallography data
The structure of Crystal Structure of N-Terminal Sars-Cov-2 NSP1 in Complex with Fragment Hit 11C6 Refined Against Anomalous Diffraction Data, PDB code: 8crm
was solved by
S.Ma,
V.Mikhailik,
N.Pinotsis,
M.W.Bowler,
F.Kozielski,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 35.63 / 1.42 |
| Space group | P 43 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 36.817, 36.817, 141.205, 90, 90, 90 |
| R / Rfree (%) | 17.9 / 20.3 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of N-Terminal Sars-Cov-2 NSP1 in Complex with Fragment Hit 11C6 Refined Against Anomalous Diffraction Data
(pdb code 8crm). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Crystal Structure of N-Terminal Sars-Cov-2 NSP1 in Complex with Fragment Hit 11C6 Refined Against Anomalous Diffraction Data, PDB code: 8crm:
In total only one binding site of Chlorine was determined in the Crystal Structure of N-Terminal Sars-Cov-2 NSP1 in Complex with Fragment Hit 11C6 Refined Against Anomalous Diffraction Data, PDB code: 8crm:
Chlorine binding site 1 out of 1 in 8crm
Go back to
Chlorine binding site 1 out
of 1 in the Crystal Structure of N-Terminal Sars-Cov-2 NSP1 in Complex with Fragment Hit 11C6 Refined Against Anomalous Diffraction Data
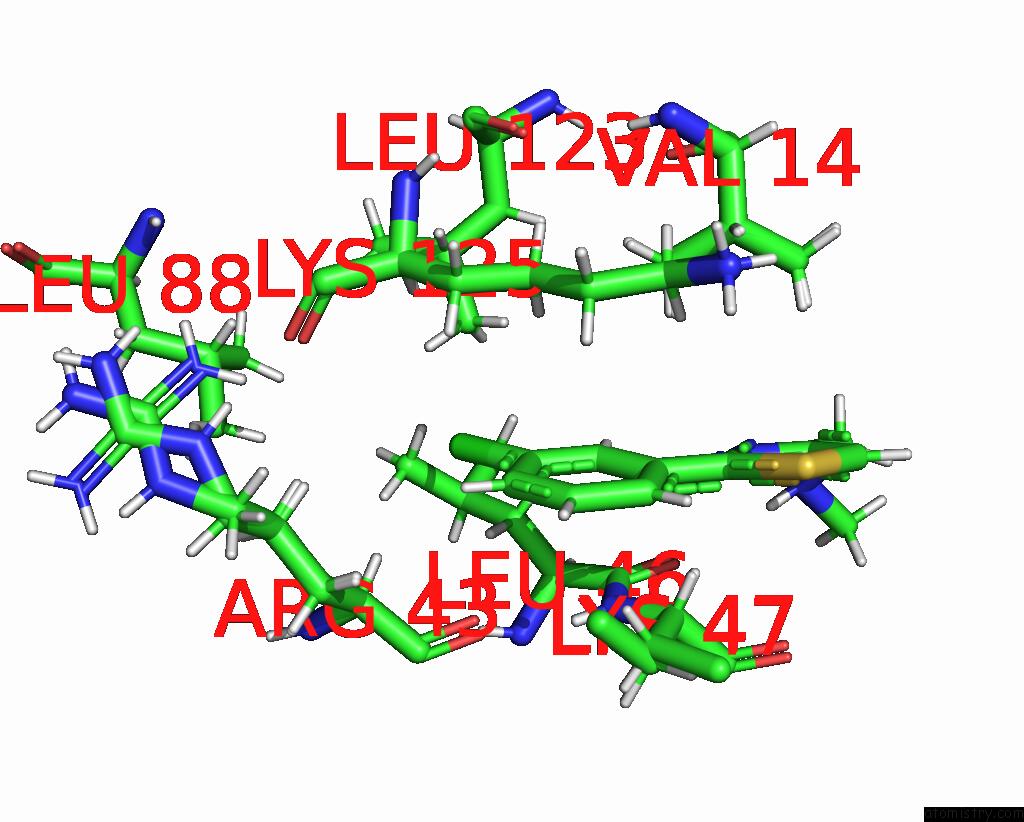
Mono view
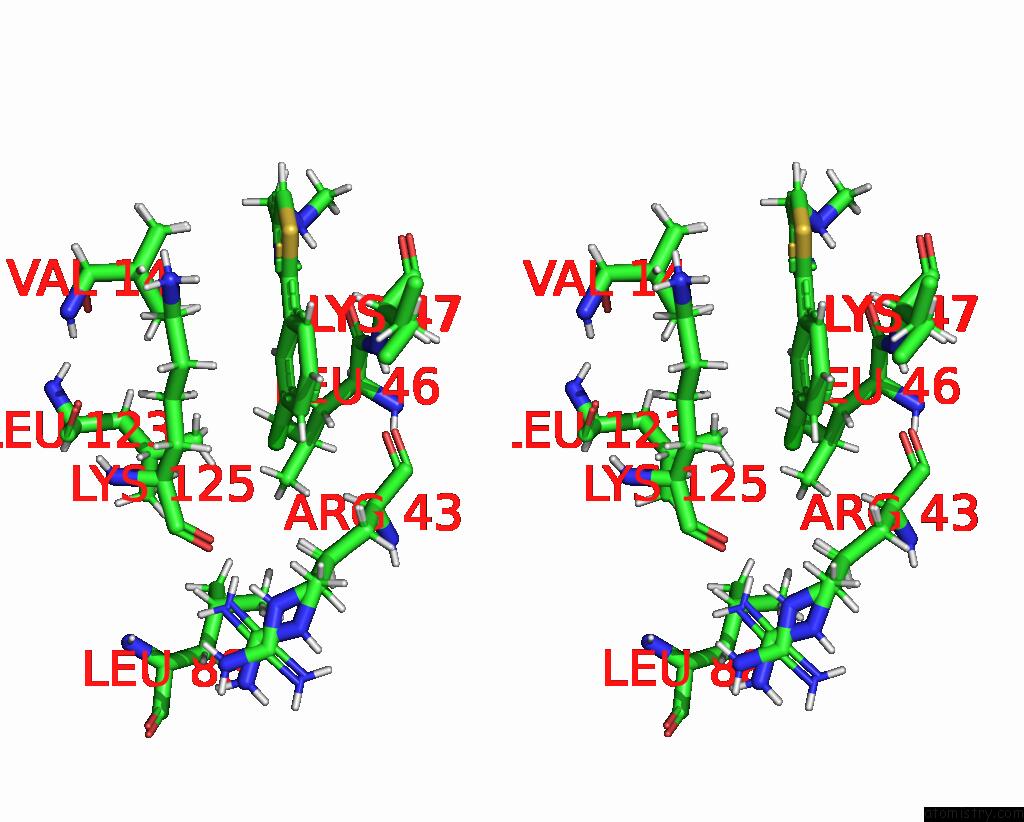
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of N-Terminal Sars-Cov-2 NSP1 in Complex with Fragment Hit 11C6 Refined Against Anomalous Diffraction Data within 5.0Å range:
|
Reference:
S.Ma,
V.Mykhaylyk,
M.W.Bowler,
N.Pinotsis,
F.Kozielski.
High-Confidence Placement of Fragments Into Electron Density Using Anomalous Diffraction-A Case Study Using Hits Targeting Sars-Cov-2 Non-Structural Protein 1. Int J Mol Sci V. 24 2023.
ISSN: ESSN 1422-0067
PubMed: 37446375
DOI: 10.3390/IJMS241311197
Page generated: Sun Jul 13 10:19:55 2025
ISSN: ESSN 1422-0067
PubMed: 37446375
DOI: 10.3390/IJMS241311197
Last articles
Na in 2WHMNa in 2WHJ
Na in 2WGD
Na in 2WGE
Na in 2WG4
Na in 2WG9
Na in 2WFT
Na in 2WFX
Na in 2WG8
Na in 2WG7