Chlorine »
PDB 8f57-8fho »
8fcb »
Chlorine in PDB 8fcb: Cryo-Em Structure of the Human TRPV4 - Rhoa in Complex with GSK1016790A
Enzymatic activity of Cryo-Em Structure of the Human TRPV4 - Rhoa in Complex with GSK1016790A
All present enzymatic activity of Cryo-Em Structure of the Human TRPV4 - Rhoa in Complex with GSK1016790A:
3.6.5.2;
3.6.5.2;
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Cryo-Em Structure of the Human TRPV4 - Rhoa in Complex with GSK1016790A
(pdb code 8fcb). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 8 binding sites of Chlorine where determined in the Cryo-Em Structure of the Human TRPV4 - Rhoa in Complex with GSK1016790A, PDB code: 8fcb:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
In total 8 binding sites of Chlorine where determined in the Cryo-Em Structure of the Human TRPV4 - Rhoa in Complex with GSK1016790A, PDB code: 8fcb:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
Chlorine binding site 1 out of 8 in 8fcb
Go back to
Chlorine binding site 1 out
of 8 in the Cryo-Em Structure of the Human TRPV4 - Rhoa in Complex with GSK1016790A
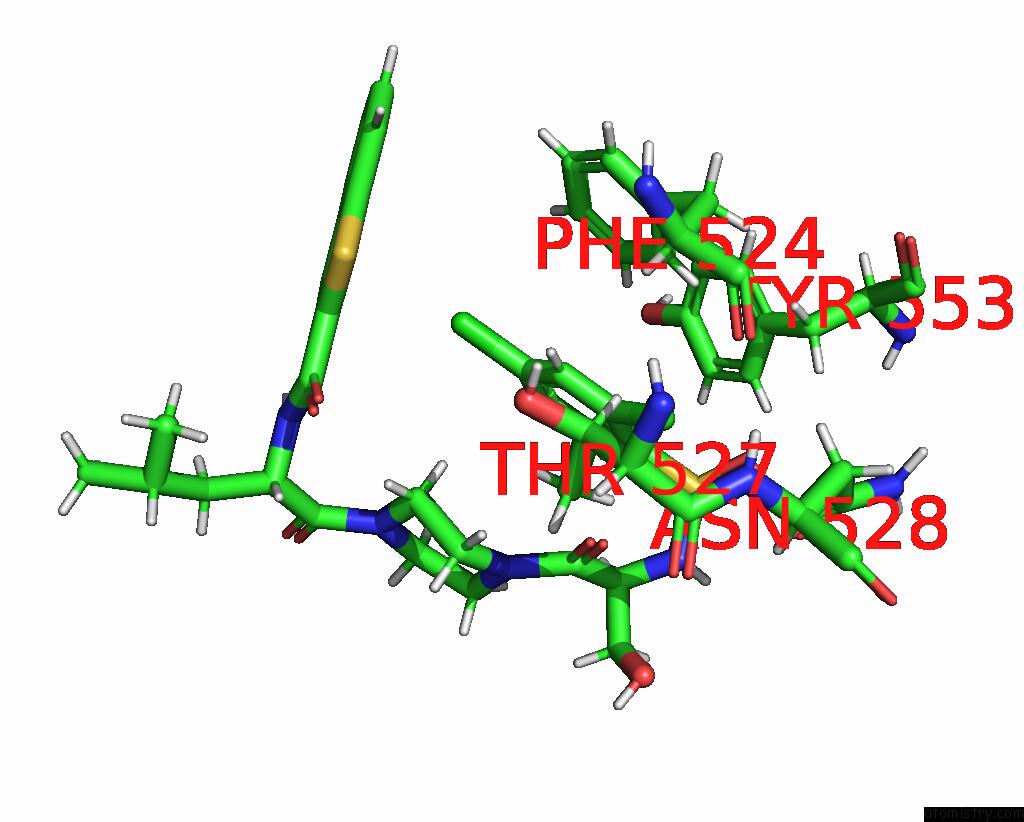
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Cryo-Em Structure of the Human TRPV4 - Rhoa in Complex with GSK1016790A within 5.0Å range:
|
Chlorine binding site 2 out of 8 in 8fcb
Go back to
Chlorine binding site 2 out
of 8 in the Cryo-Em Structure of the Human TRPV4 - Rhoa in Complex with GSK1016790A
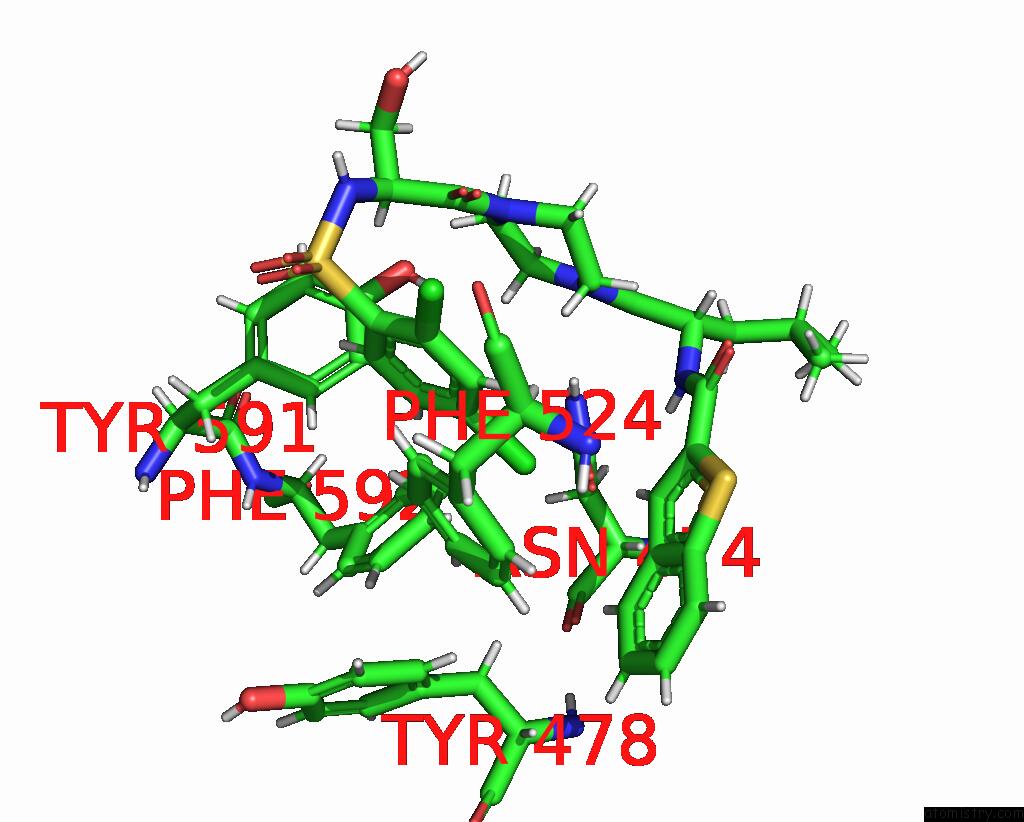
Mono view
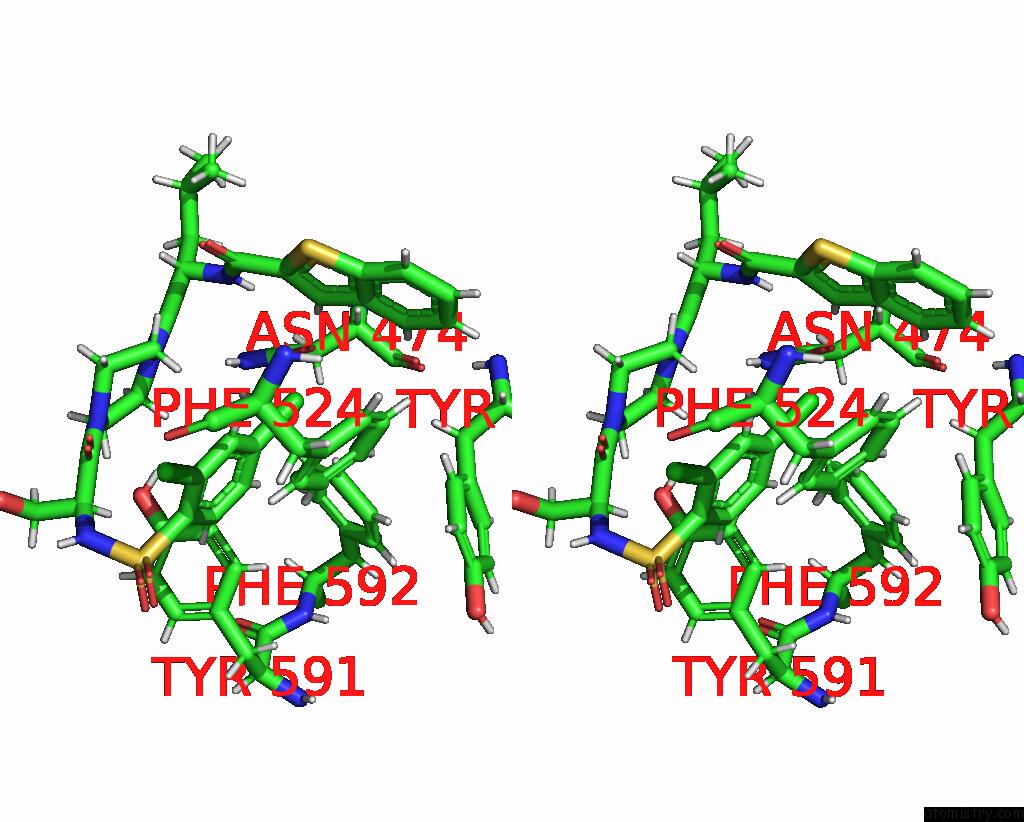
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Cryo-Em Structure of the Human TRPV4 - Rhoa in Complex with GSK1016790A within 5.0Å range:
|
Chlorine binding site 3 out of 8 in 8fcb
Go back to
Chlorine binding site 3 out
of 8 in the Cryo-Em Structure of the Human TRPV4 - Rhoa in Complex with GSK1016790A
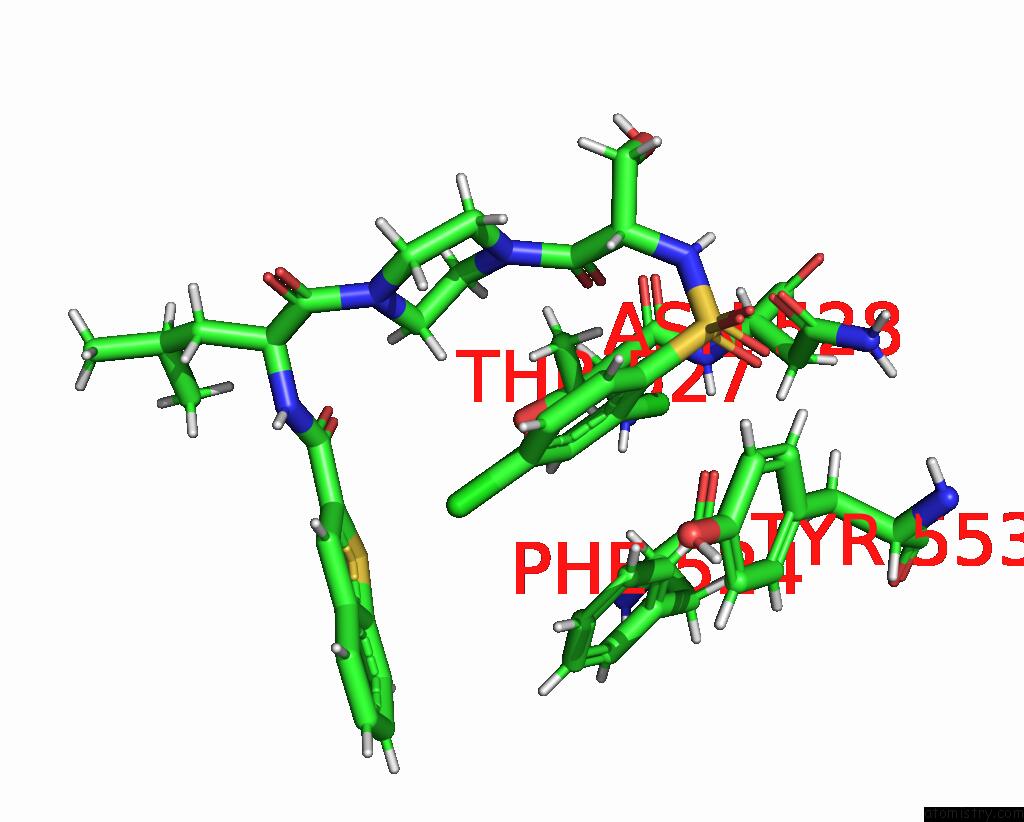
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Cryo-Em Structure of the Human TRPV4 - Rhoa in Complex with GSK1016790A within 5.0Å range:
|
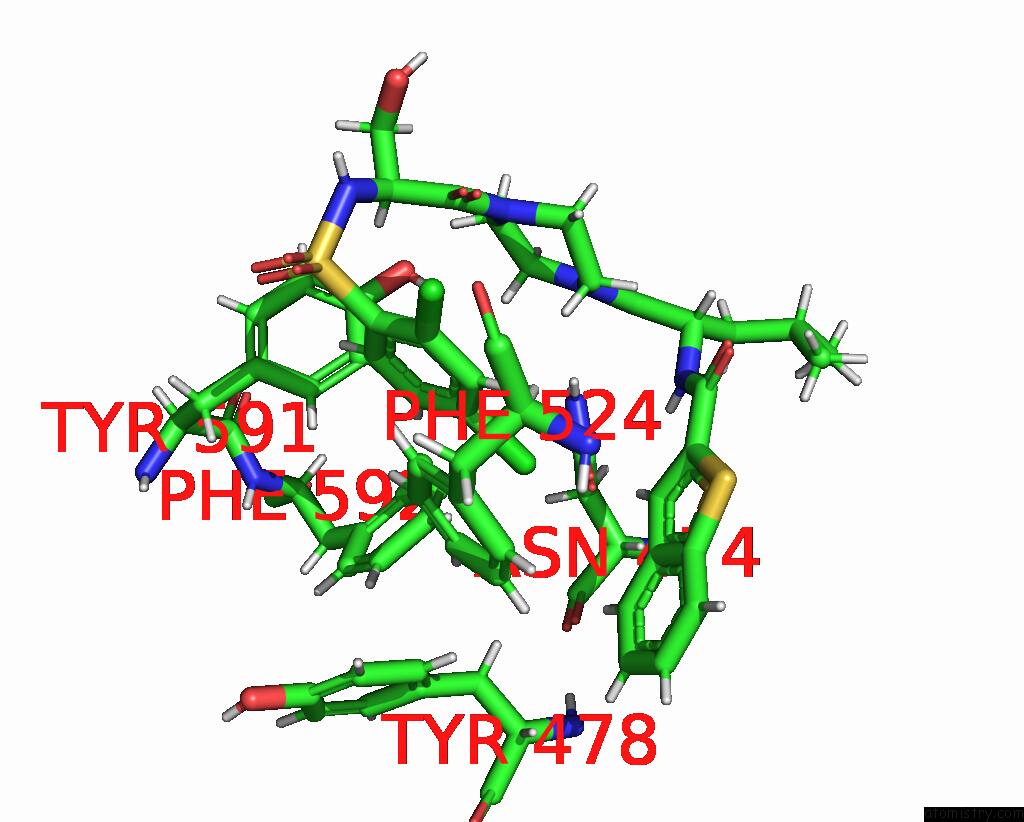
Chlorine binding site 4 out of 8 in 8fcb
Go back to
Chlorine binding site 4 out
of 8 in the Cryo-Em Structure of the Human TRPV4 - Rhoa in Complex with GSK1016790A
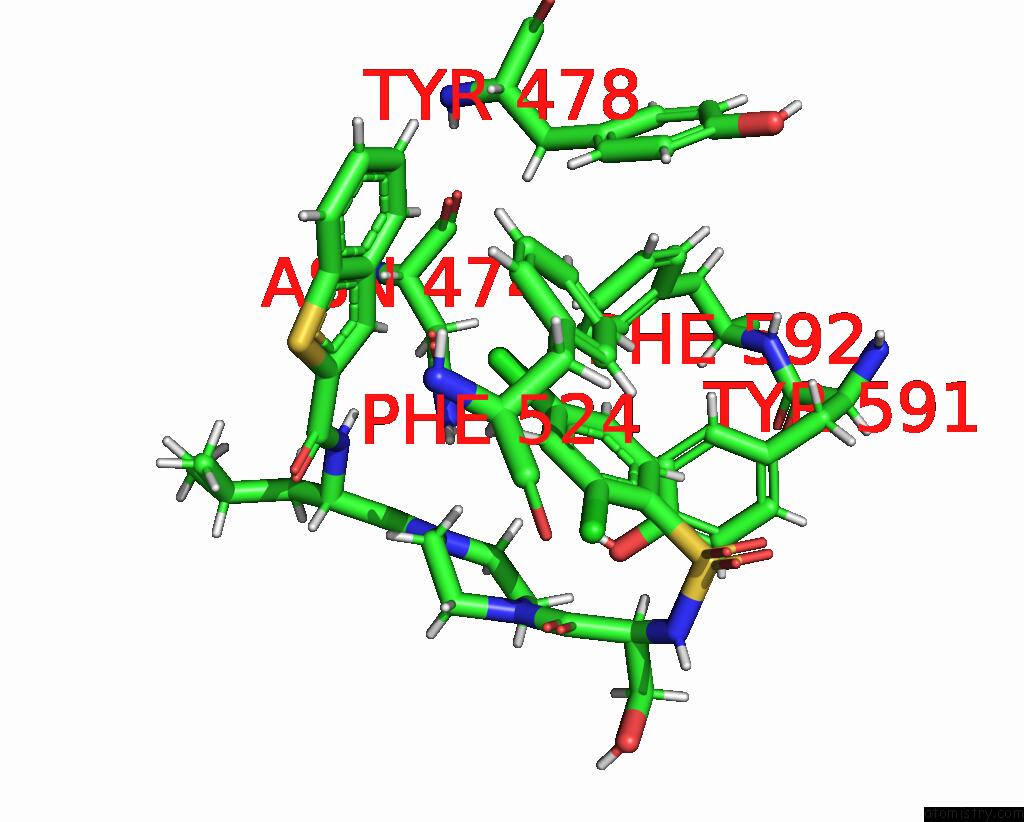
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Cryo-Em Structure of the Human TRPV4 - Rhoa in Complex with GSK1016790A within 5.0Å range:
|
Chlorine binding site 5 out of 8 in 8fcb
Go back to
Chlorine binding site 5 out
of 8 in the Cryo-Em Structure of the Human TRPV4 - Rhoa in Complex with GSK1016790A
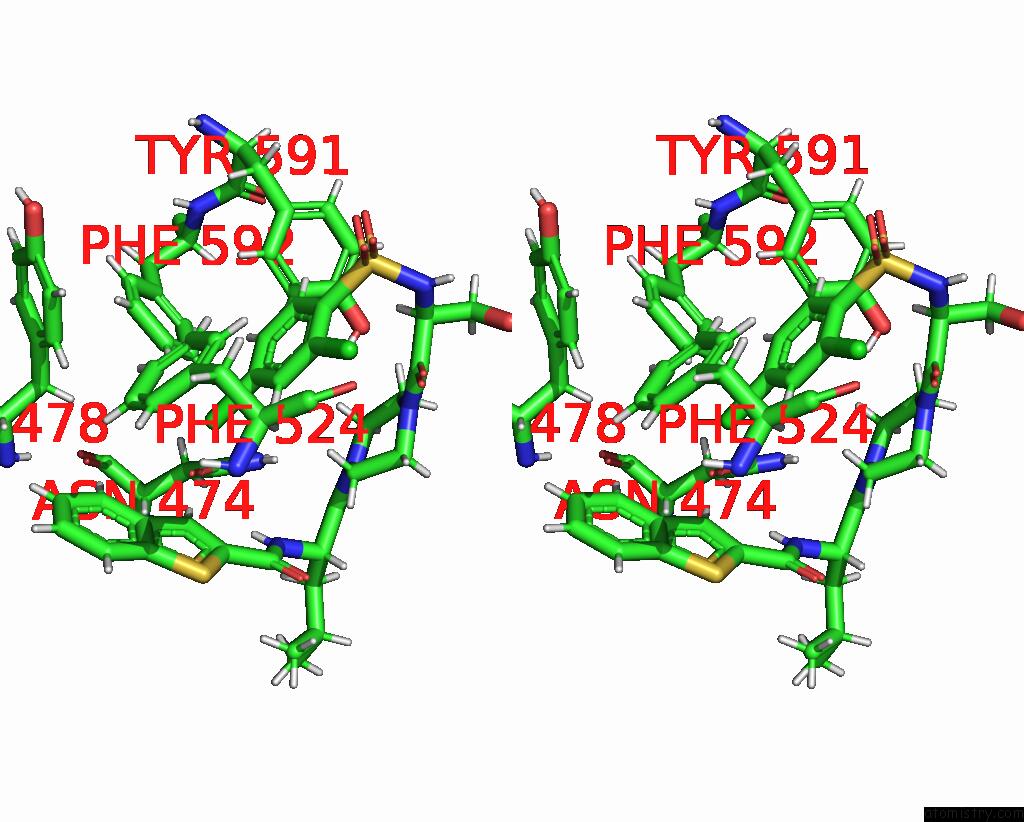
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Cryo-Em Structure of the Human TRPV4 - Rhoa in Complex with GSK1016790A within 5.0Å range:
|
Chlorine binding site 6 out of 8 in 8fcb
Go back to
Chlorine binding site 6 out
of 8 in the Cryo-Em Structure of the Human TRPV4 - Rhoa in Complex with GSK1016790A
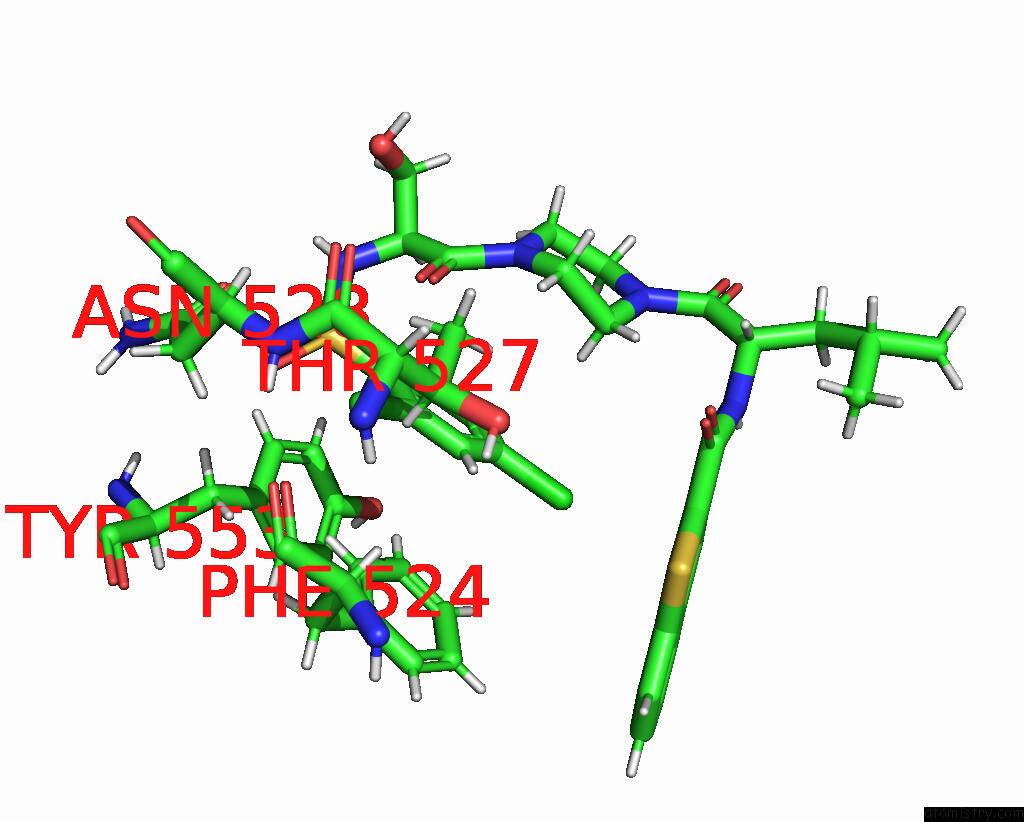
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of Cryo-Em Structure of the Human TRPV4 - Rhoa in Complex with GSK1016790A within 5.0Å range:
|
Chlorine binding site 7 out of 8 in 8fcb
Go back to
Chlorine binding site 7 out
of 8 in the Cryo-Em Structure of the Human TRPV4 - Rhoa in Complex with GSK1016790A
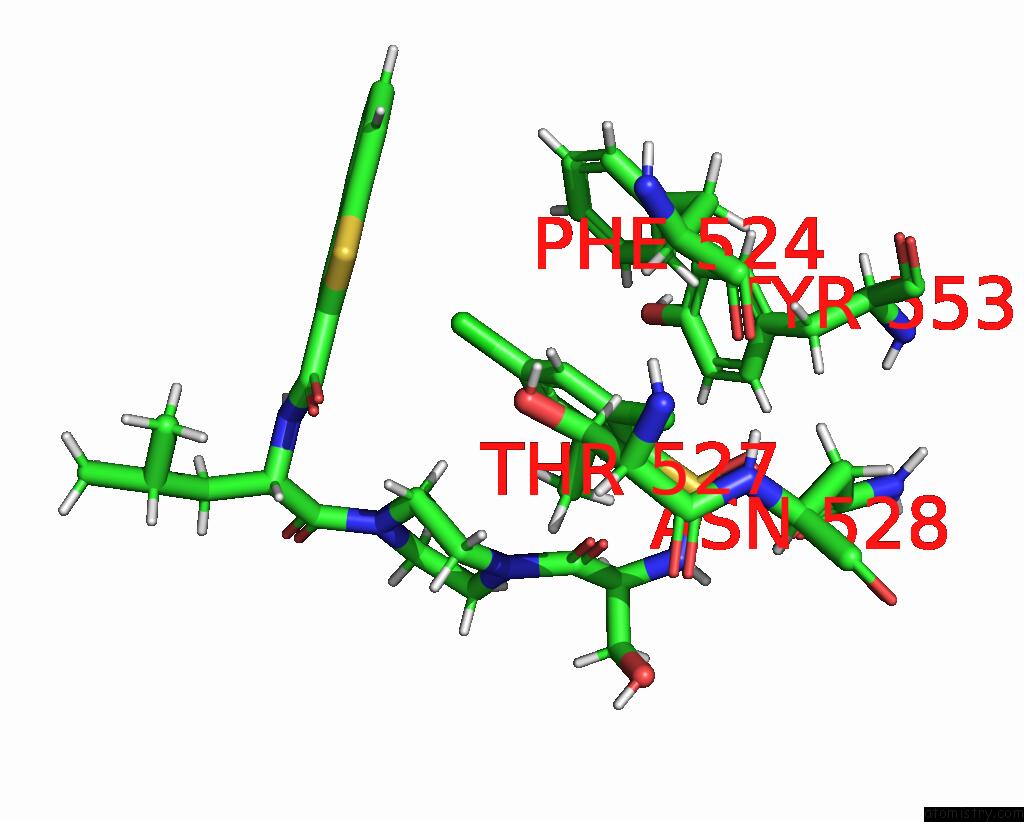
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 7 of Cryo-Em Structure of the Human TRPV4 - Rhoa in Complex with GSK1016790A within 5.0Å range:
|
Chlorine binding site 8 out of 8 in 8fcb
Go back to
Chlorine binding site 8 out
of 8 in the Cryo-Em Structure of the Human TRPV4 - Rhoa in Complex with GSK1016790A

Mono view
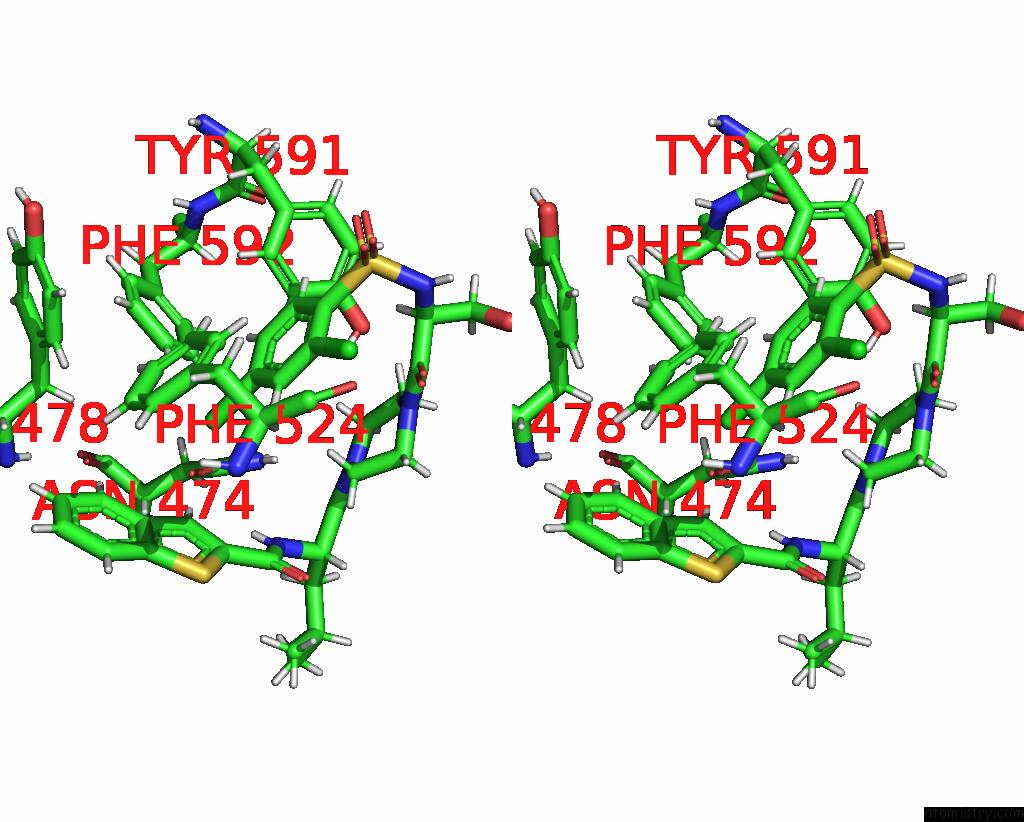
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 8 of Cryo-Em Structure of the Human TRPV4 - Rhoa in Complex with GSK1016790A within 5.0Å range:
|
Reference:
D.H.Kwon,
F.Zhang,
B.A.Mccray,
S.Feng,
M.Kumar,
J.M.Sullivan,
W.Im,
C.J.Sumner,
S.Y.Lee.
TRPV4-Rho Gtpase Complex Structures Reveal Mechanisms of Gating and Disease. Nat Commun V. 14 3732 2023.
ISSN: ESSN 2041-1723
PubMed: 37353484
DOI: 10.1038/S41467-023-39345-0
Page generated: Sun Jul 13 11:21:13 2025
ISSN: ESSN 2041-1723
PubMed: 37353484
DOI: 10.1038/S41467-023-39345-0
Last articles
K in 6T2GK in 6SUU
K in 6T0I
K in 6S6Y
K in 6SWR
K in 6SSS
K in 6SLS
K in 6SJ9
K in 6SJ6
K in 6S3K