Chlorine »
PDB 8ii0-8it8 »
8iq0 »
Chlorine in PDB 8iq0: Crystal Structure of Hydrogen Sulfide-Bound Superoxide Dismutase in Oxidized State
Enzymatic activity of Crystal Structure of Hydrogen Sulfide-Bound Superoxide Dismutase in Oxidized State
All present enzymatic activity of Crystal Structure of Hydrogen Sulfide-Bound Superoxide Dismutase in Oxidized State:
1.15.1.1;
1.15.1.1;
Protein crystallography data
The structure of Crystal Structure of Hydrogen Sulfide-Bound Superoxide Dismutase in Oxidized State, PDB code: 8iq0
was solved by
J.H.Zhou,
W.X.Huang,
R.X.Cheng,
P.J.Zhang,
Y.C.Zhu,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 30.68 / 1.88 |
| Space group | P 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 74.348, 91.474, 91.164, 85, 78.06, 66.76 |
| R / Rfree (%) | 20.1 / 24.5 |
Other elements in 8iq0:
The structure of Crystal Structure of Hydrogen Sulfide-Bound Superoxide Dismutase in Oxidized State also contains other interesting chemical elements:
| Copper | (Cu) | 19 atoms |
| Zinc | (Zn) | 16 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of Hydrogen Sulfide-Bound Superoxide Dismutase in Oxidized State
(pdb code 8iq0). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Crystal Structure of Hydrogen Sulfide-Bound Superoxide Dismutase in Oxidized State, PDB code: 8iq0:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Crystal Structure of Hydrogen Sulfide-Bound Superoxide Dismutase in Oxidized State, PDB code: 8iq0:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 8iq0
Go back to
Chlorine binding site 1 out
of 2 in the Crystal Structure of Hydrogen Sulfide-Bound Superoxide Dismutase in Oxidized State
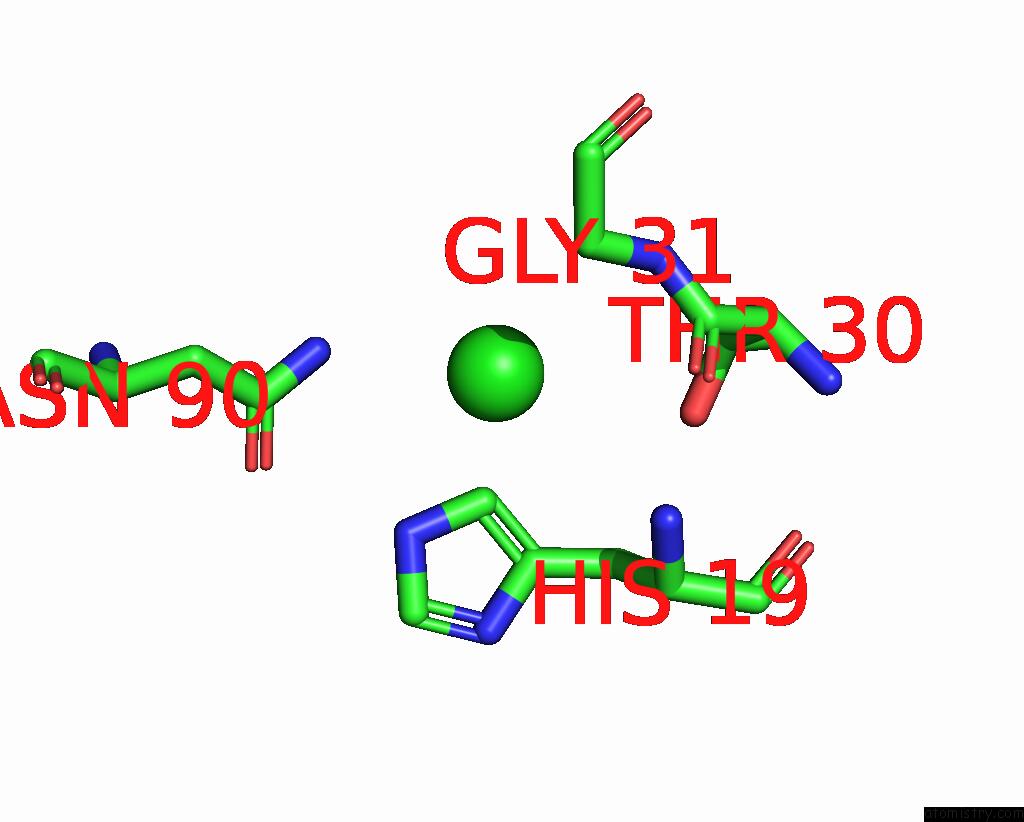
Mono view
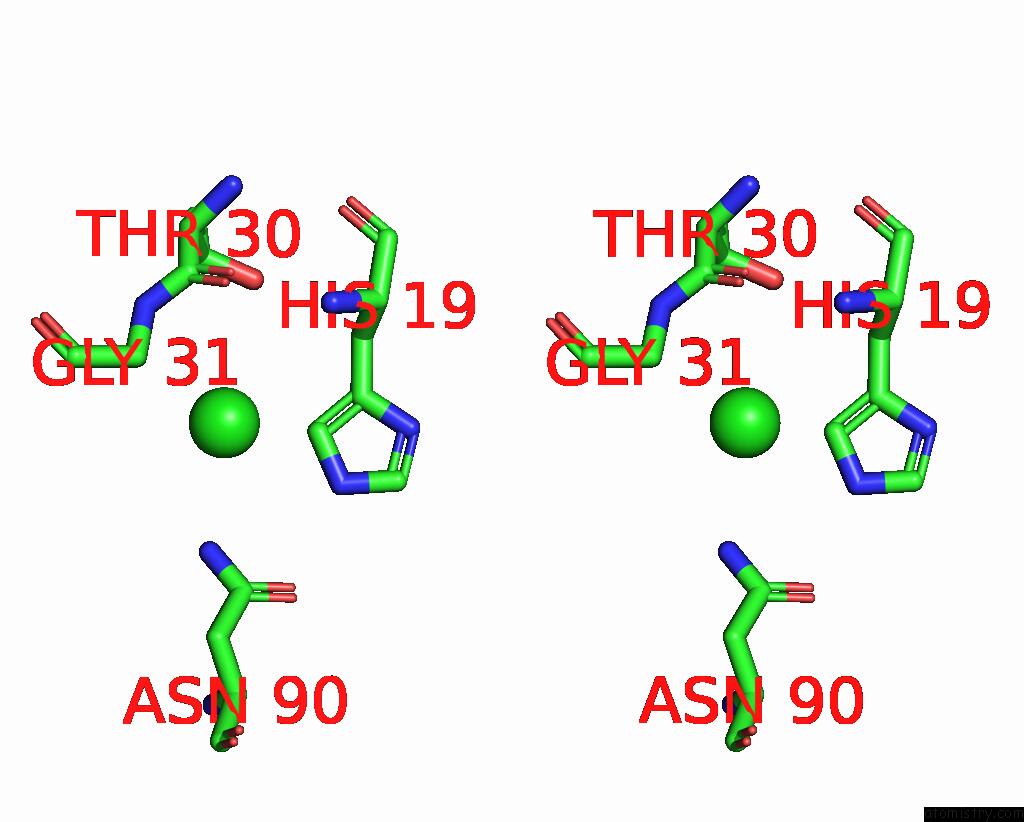
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Hydrogen Sulfide-Bound Superoxide Dismutase in Oxidized State within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 8iq0
Go back to
Chlorine binding site 2 out
of 2 in the Crystal Structure of Hydrogen Sulfide-Bound Superoxide Dismutase in Oxidized State
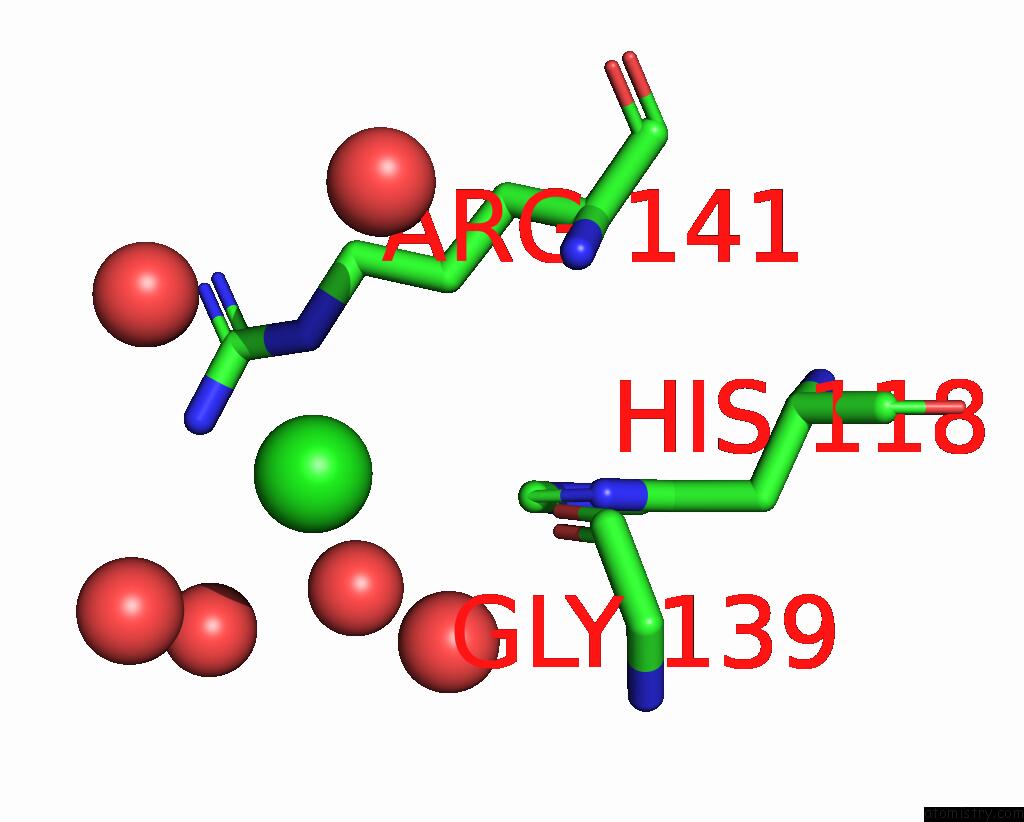
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of Hydrogen Sulfide-Bound Superoxide Dismutase in Oxidized State within 5.0Å range:
|
Reference:
D.D.Wu,
S.Jin,
R.X.Cheng,
W.J.Cai,
W.L.Xue,
Q.Q.Zhang,
L.J.Yang,
Q.Zhu,
M.Y.Li,
G.Lin,
Y.Z.Wang,
X.P.Mu,
Y.Wang,
I.Y.Zhang,
Q.Zhang,
Y.Chen,
S.Y.Cai,
B.Tan,
Y.Li,
Y.Q.Chen,
P.J.Zhang,
C.Sun,
Y.Yin,
M.J.Wang,
Y.Z.Zhu,
B.B.Tao,
J.H.Zhou,
W.X.Huang,
Y.C.Zhu.
Hydrogen Sulfide Functions As A Micro-Modulator Bound at the Copper Active Site of Cu/Zn-Sod to Regulate the Catalytic Activity of the Enzyme. Cell Rep V. 42 12750 2023.
ISSN: ESSN 2211-1247
PubMed: 37421623
DOI: 10.1016/J.CELREP.2023.112750
Page generated: Sun Jul 13 12:16:08 2025
ISSN: ESSN 2211-1247
PubMed: 37421623
DOI: 10.1016/J.CELREP.2023.112750
Last articles
K in 5E33K in 5E2Q
K in 5E1A
K in 5DWX
K in 5DUN
K in 5DSX
K in 5DRY
K in 5DRT
K in 5DMJ
K in 5DQK