Chlorine »
PDB 8itb-8j4w »
8j0j »
Chlorine in PDB 8j0j: ATSLAC1 8D Mutant in Closed State
Chlorine Binding Sites:
The binding sites of Chlorine atom in the ATSLAC1 8D Mutant in Closed State
(pdb code 8j0j). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 3 binding sites of Chlorine where determined in the ATSLAC1 8D Mutant in Closed State, PDB code: 8j0j:
Jump to Chlorine binding site number: 1; 2; 3;
In total 3 binding sites of Chlorine where determined in the ATSLAC1 8D Mutant in Closed State, PDB code: 8j0j:
Jump to Chlorine binding site number: 1; 2; 3;
Chlorine binding site 1 out of 3 in 8j0j
Go back to
Chlorine binding site 1 out
of 3 in the ATSLAC1 8D Mutant in Closed State
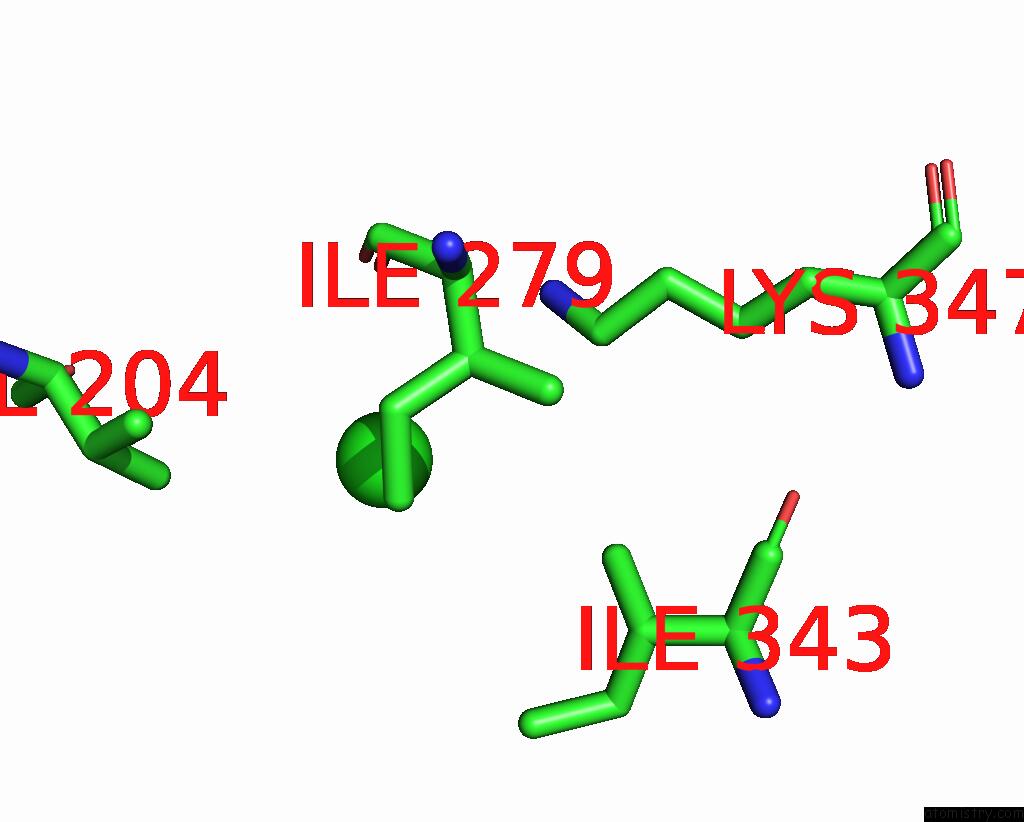
Mono view
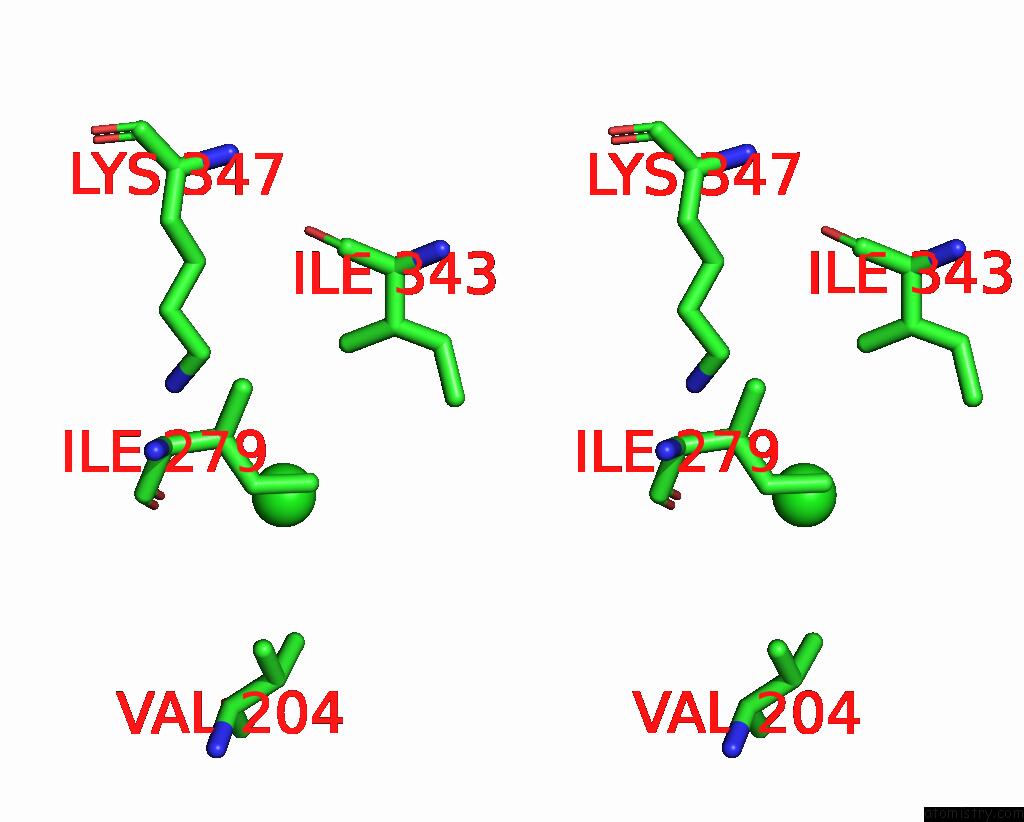
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of ATSLAC1 8D Mutant in Closed State within 5.0Å range:
|
Chlorine binding site 2 out of 3 in 8j0j
Go back to
Chlorine binding site 2 out
of 3 in the ATSLAC1 8D Mutant in Closed State
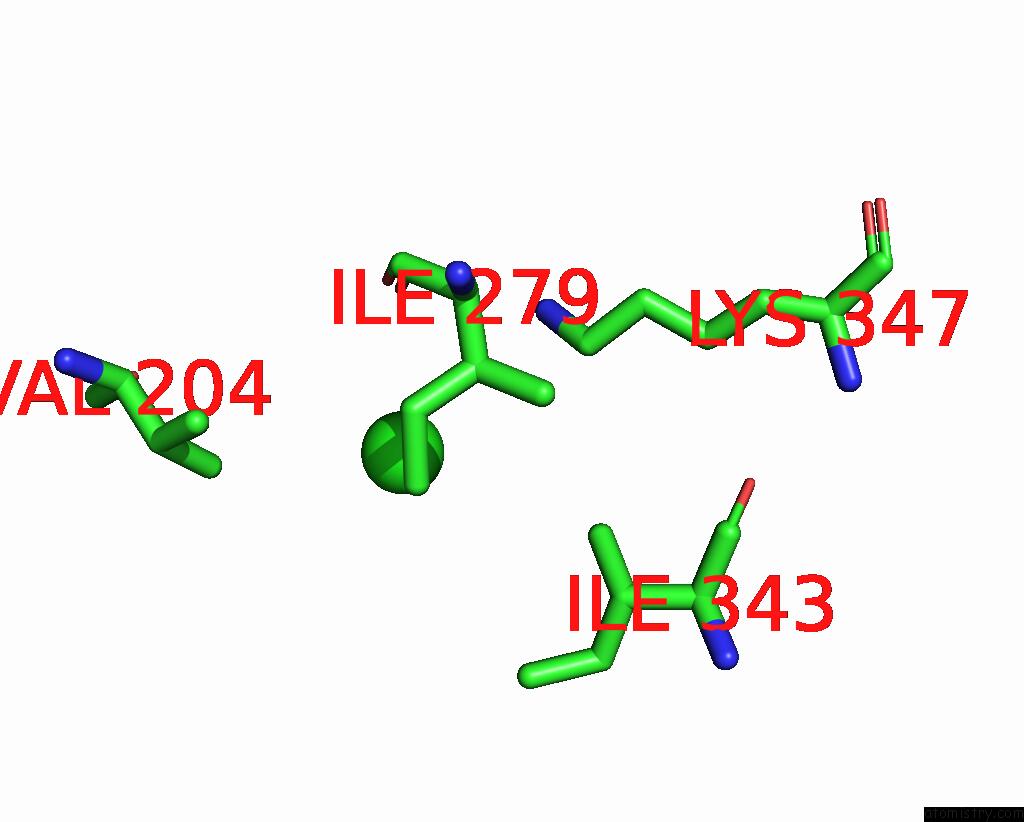
Mono view
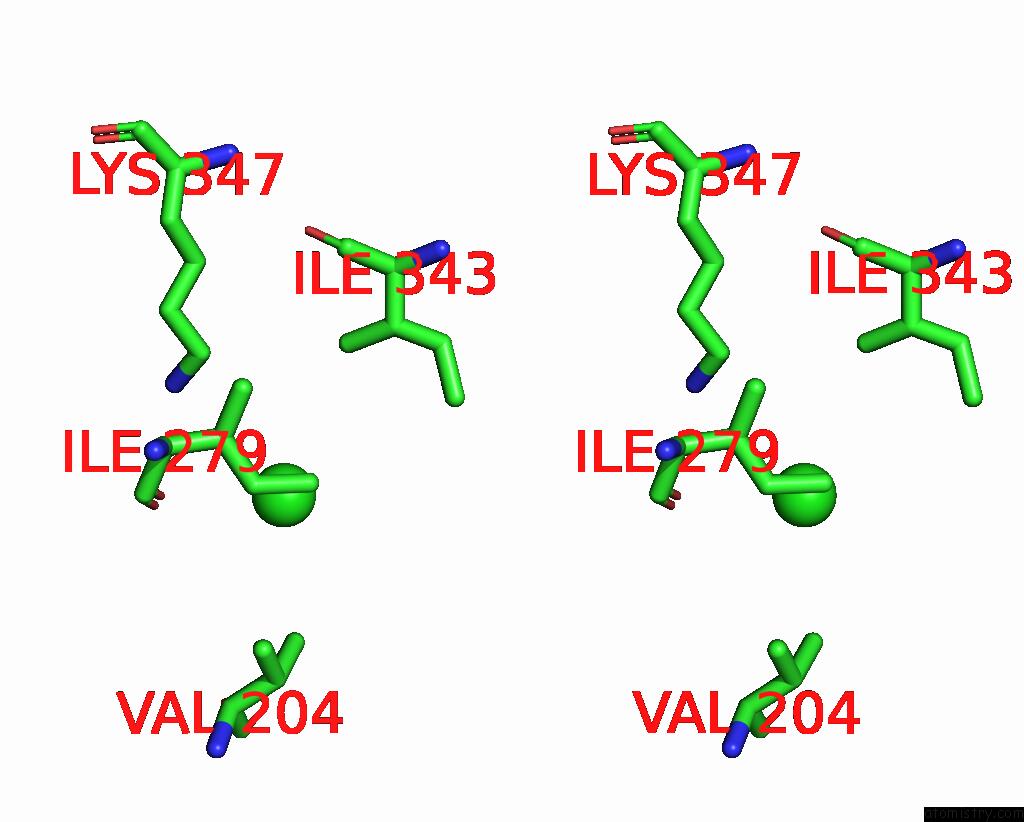
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of ATSLAC1 8D Mutant in Closed State within 5.0Å range:
|
Chlorine binding site 3 out of 3 in 8j0j
Go back to
Chlorine binding site 3 out
of 3 in the ATSLAC1 8D Mutant in Closed State
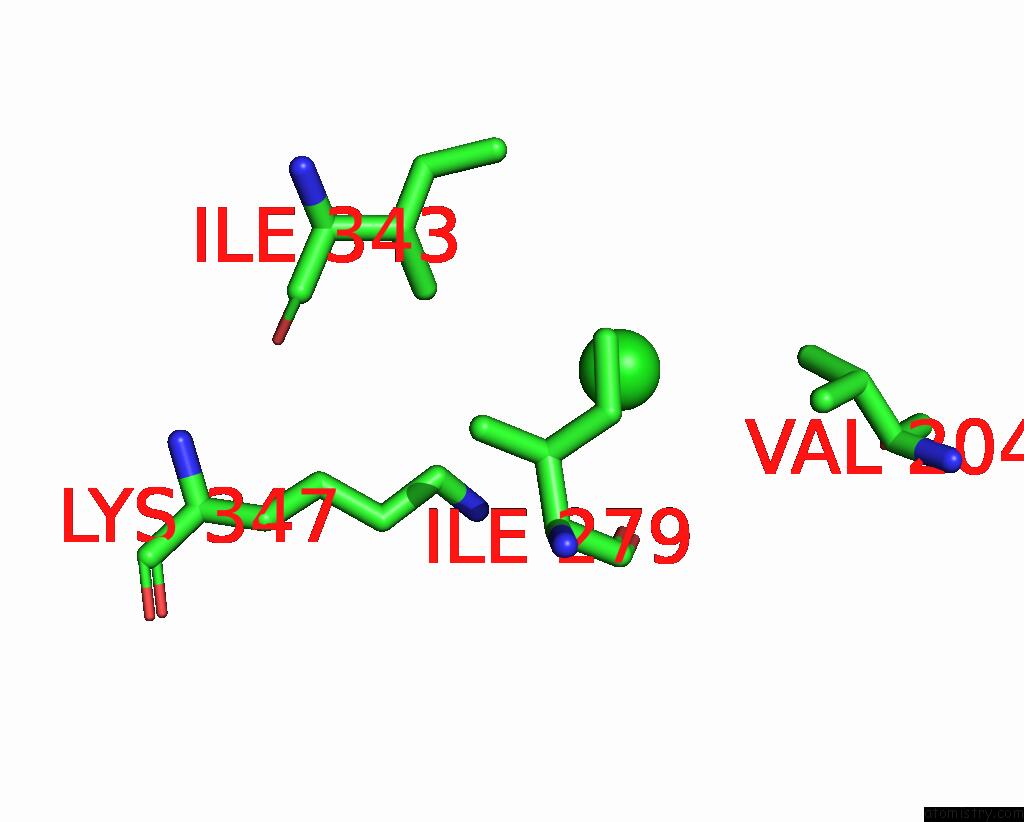
Mono view
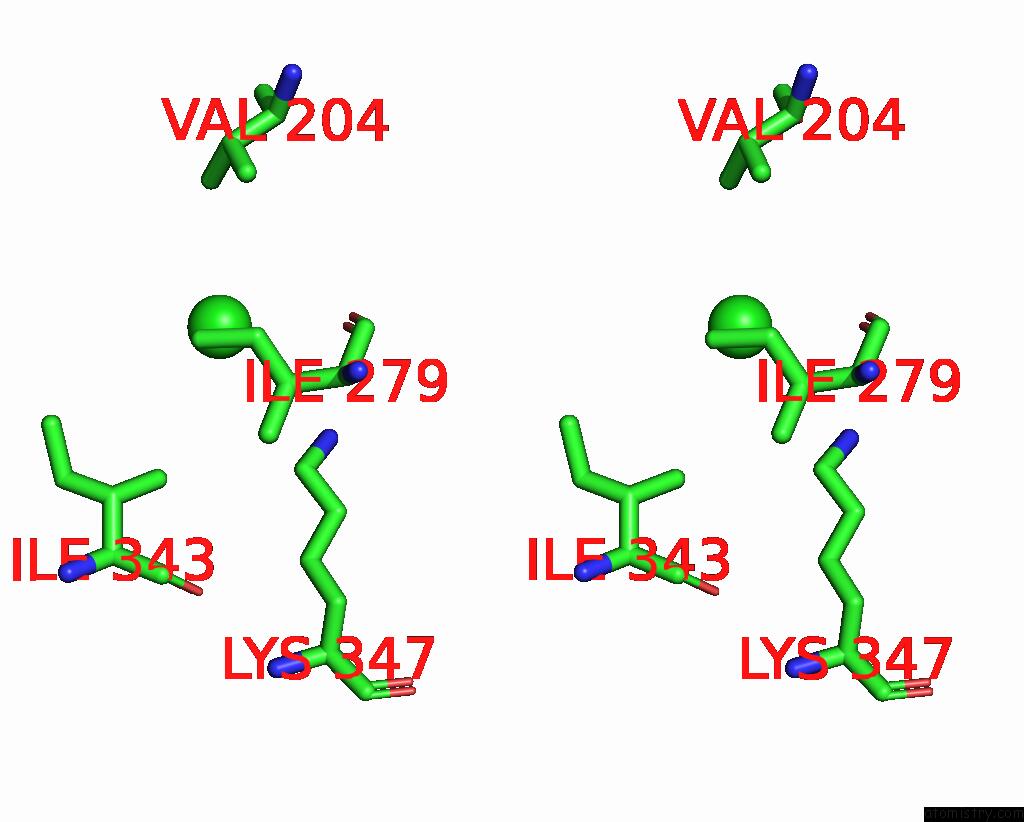
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of ATSLAC1 8D Mutant in Closed State within 5.0Å range:
|
Reference:
Y.Lee,
S.Lee.
Cryo-Em Structures of the Plant Anion Channel SLAC1 From Arabidopsis Thaliana Suggest A Combined Activation Model To Be Published 2023.
DOI: 10.1038/S41467-023-43193-3
Page generated: Sun Jul 13 12:32:36 2025
DOI: 10.1038/S41467-023-43193-3
Last articles
Mn in 4EE1Mn in 4EDV
Mn in 4EDT
Mn in 4EDR
Mn in 4EAY
Mn in 4EDK
Mn in 4EDG
Mn in 4E5L
Mn in 4EAC
Mn in 4E7L