Chlorine »
PDB 8oqd-8p0m »
8ox3 »
Chlorine in PDB 8ox3: Structure of the Murine Lyve-1 (Lymphatic Vessel Endothelial Receptor- 1) Hyaluronan Binding Domain Bound with Octasaccharide Hyaluronan.
Protein crystallography data
The structure of Structure of the Murine Lyve-1 (Lymphatic Vessel Endothelial Receptor- 1) Hyaluronan Binding Domain Bound with Octasaccharide Hyaluronan., PDB code: 8ox3
was solved by
T.Ni,
N.Bannerji,
D.J.Jackson,
R.J.C.Gilbert,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 46.90 / 1.05 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 86.107, 56.963, 30.908, 90, 106.27, 90 |
| R / Rfree (%) | 15.1 / 16.3 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Structure of the Murine Lyve-1 (Lymphatic Vessel Endothelial Receptor- 1) Hyaluronan Binding Domain Bound with Octasaccharide Hyaluronan.
(pdb code 8ox3). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Structure of the Murine Lyve-1 (Lymphatic Vessel Endothelial Receptor- 1) Hyaluronan Binding Domain Bound with Octasaccharide Hyaluronan., PDB code: 8ox3:
In total only one binding site of Chlorine was determined in the Structure of the Murine Lyve-1 (Lymphatic Vessel Endothelial Receptor- 1) Hyaluronan Binding Domain Bound with Octasaccharide Hyaluronan., PDB code: 8ox3:
Chlorine binding site 1 out of 1 in 8ox3
Go back to
Chlorine binding site 1 out
of 1 in the Structure of the Murine Lyve-1 (Lymphatic Vessel Endothelial Receptor- 1) Hyaluronan Binding Domain Bound with Octasaccharide Hyaluronan.
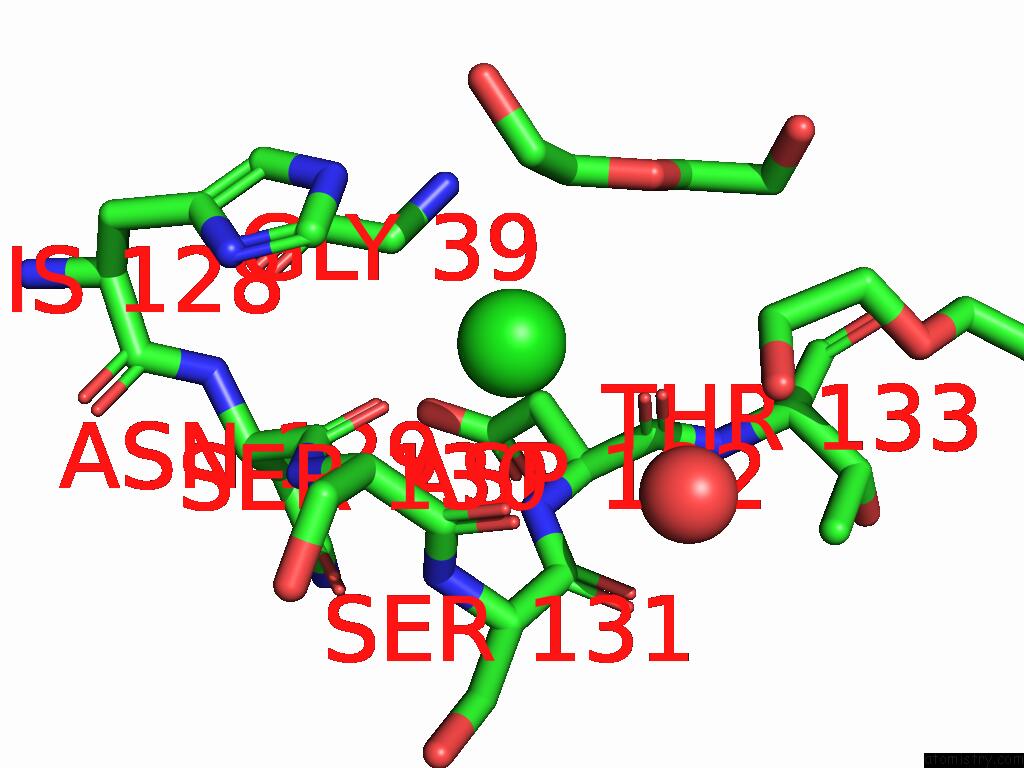
Mono view
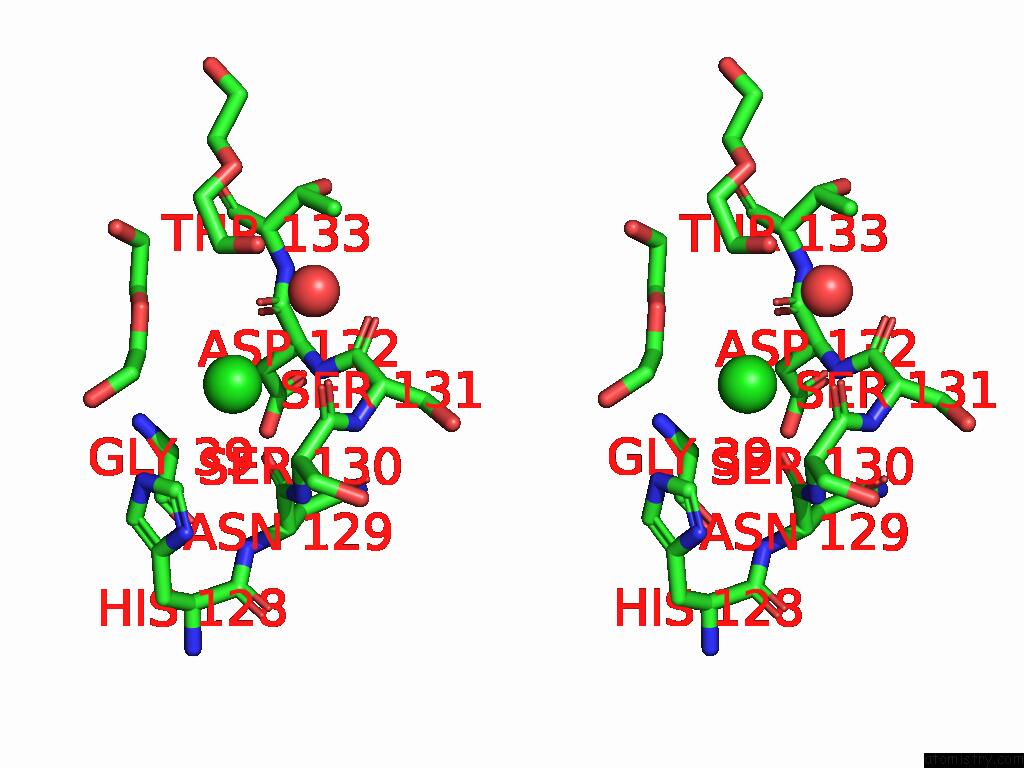
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Structure of the Murine Lyve-1 (Lymphatic Vessel Endothelial Receptor- 1) Hyaluronan Binding Domain Bound with Octasaccharide Hyaluronan. within 5.0Å range:
|
Reference:
F.Bano,
S.Bannerji,
T.Ni,
D.E.Green,
P.L.Deangelis,
E.Paci,
M.Lepsik,
R.J.C.Gilbert,
R.P.Richter,
D.J.Jackson.
A Novel Sliding Interaction with Hyaluronan Underlies the Mode of Action of Lyve-1, the Receptor For Leucocyte Entry and Trafficking in the Lymphatics To Be Published.
Page generated: Sun Jul 13 12:57:16 2025
Last articles
I in 9ET8I in 9ET1
I in 9ESZ
I in 9E56
I in 9DXP
I in 9E4X
I in 9E50
I in 9DQ1
I in 9DWY
I in 9DAJ