Chlorine »
PDB 8qtl-8r5v »
8qzt »
Chlorine in PDB 8qzt: Single Particle Cryo-Em Co-Structure of E. Coli Acrb with Bound BDM91531 Inhibitor at 3.52 A Resolution
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Single Particle Cryo-Em Co-Structure of E. Coli Acrb with Bound BDM91531 Inhibitor at 3.52 A Resolution
(pdb code 8qzt). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Single Particle Cryo-Em Co-Structure of E. Coli Acrb with Bound BDM91531 Inhibitor at 3.52 A Resolution, PDB code: 8qzt:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Single Particle Cryo-Em Co-Structure of E. Coli Acrb with Bound BDM91531 Inhibitor at 3.52 A Resolution, PDB code: 8qzt:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 8qzt
Go back to
Chlorine binding site 1 out
of 2 in the Single Particle Cryo-Em Co-Structure of E. Coli Acrb with Bound BDM91531 Inhibitor at 3.52 A Resolution
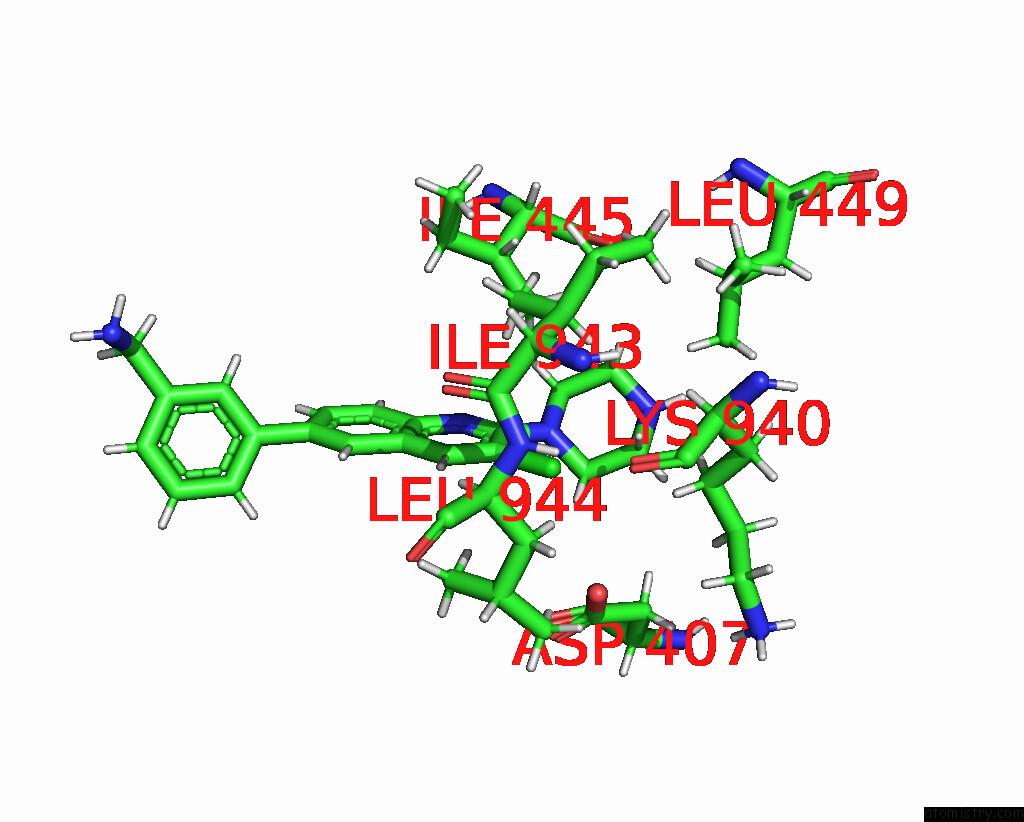
Mono view
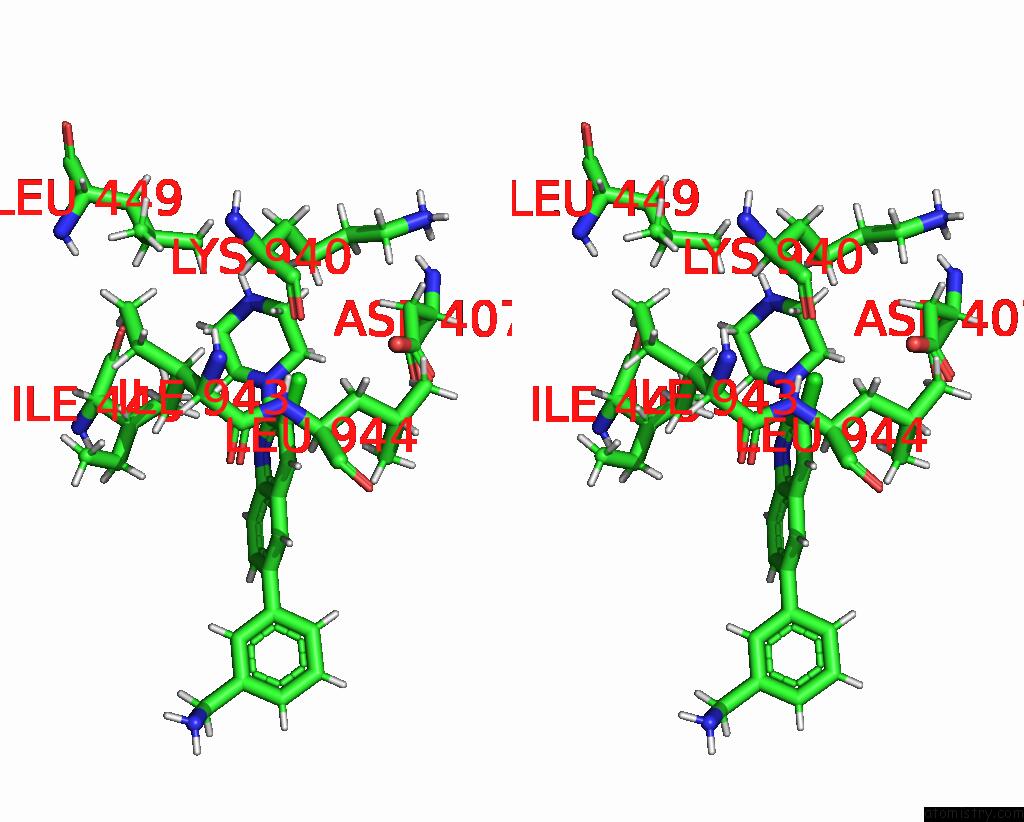
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Single Particle Cryo-Em Co-Structure of E. Coli Acrb with Bound BDM91531 Inhibitor at 3.52 A Resolution within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 8qzt
Go back to
Chlorine binding site 2 out
of 2 in the Single Particle Cryo-Em Co-Structure of E. Coli Acrb with Bound BDM91531 Inhibitor at 3.52 A Resolution
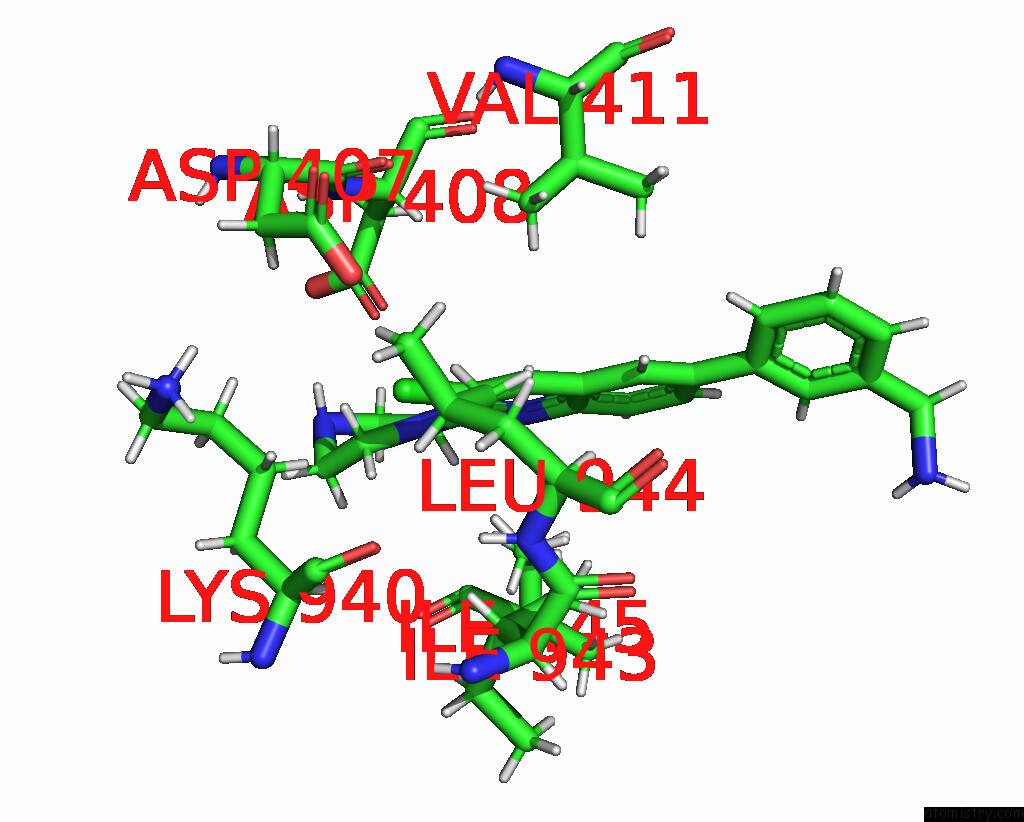
Mono view
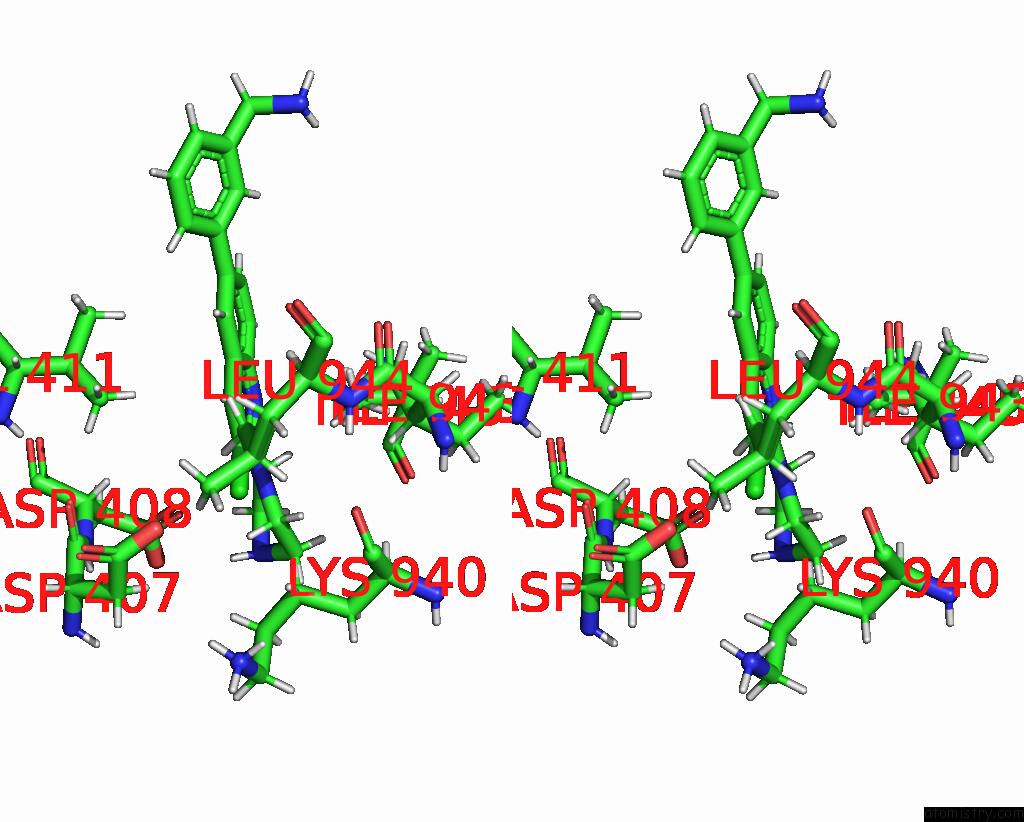
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Single Particle Cryo-Em Co-Structure of E. Coli Acrb with Bound BDM91531 Inhibitor at 3.52 A Resolution within 5.0Å range:
|
Reference:
C.Boernsen,
R.T.Mueller.
Single Particle Cryo-Em Co-Structure of E. Coli Acrb with Bound BDM91531 Inhibitor at 3.52 A Resolution To Be Published.
Page generated: Sun Jul 13 13:42:22 2025
Last articles
Mn in 4F7IMn in 4FBK
Mn in 4F6E
Mn in 4F5R
Mn in 4EWT
Mn in 4F5Q
Mn in 4F3O
Mn in 4F2D
Mn in 4F4K
Mn in 4F2S