Chlorine »
PDB 8rj2-8rwz »
8rpa »
Chlorine in PDB 8rpa: Crystal Structure of Zea Mays Adenosine Kinase 3 (ZMADK3) in Complex with AP5A
Enzymatic activity of Crystal Structure of Zea Mays Adenosine Kinase 3 (ZMADK3) in Complex with AP5A
All present enzymatic activity of Crystal Structure of Zea Mays Adenosine Kinase 3 (ZMADK3) in Complex with AP5A:
2.7.1.20;
2.7.1.20;
Protein crystallography data
The structure of Crystal Structure of Zea Mays Adenosine Kinase 3 (ZMADK3) in Complex with AP5A, PDB code: 8rpa
was solved by
S.Morera,
D.Kopecny,
A.Vigouroux,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 47.95 / 2.26 |
| Space group | I 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 51.049, 117.49, 165.958, 90, 90, 90 |
| R / Rfree (%) | 20.8 / 25.7 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of Zea Mays Adenosine Kinase 3 (ZMADK3) in Complex with AP5A
(pdb code 8rpa). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Crystal Structure of Zea Mays Adenosine Kinase 3 (ZMADK3) in Complex with AP5A, PDB code: 8rpa:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Crystal Structure of Zea Mays Adenosine Kinase 3 (ZMADK3) in Complex with AP5A, PDB code: 8rpa:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 8rpa
Go back to
Chlorine binding site 1 out
of 2 in the Crystal Structure of Zea Mays Adenosine Kinase 3 (ZMADK3) in Complex with AP5A
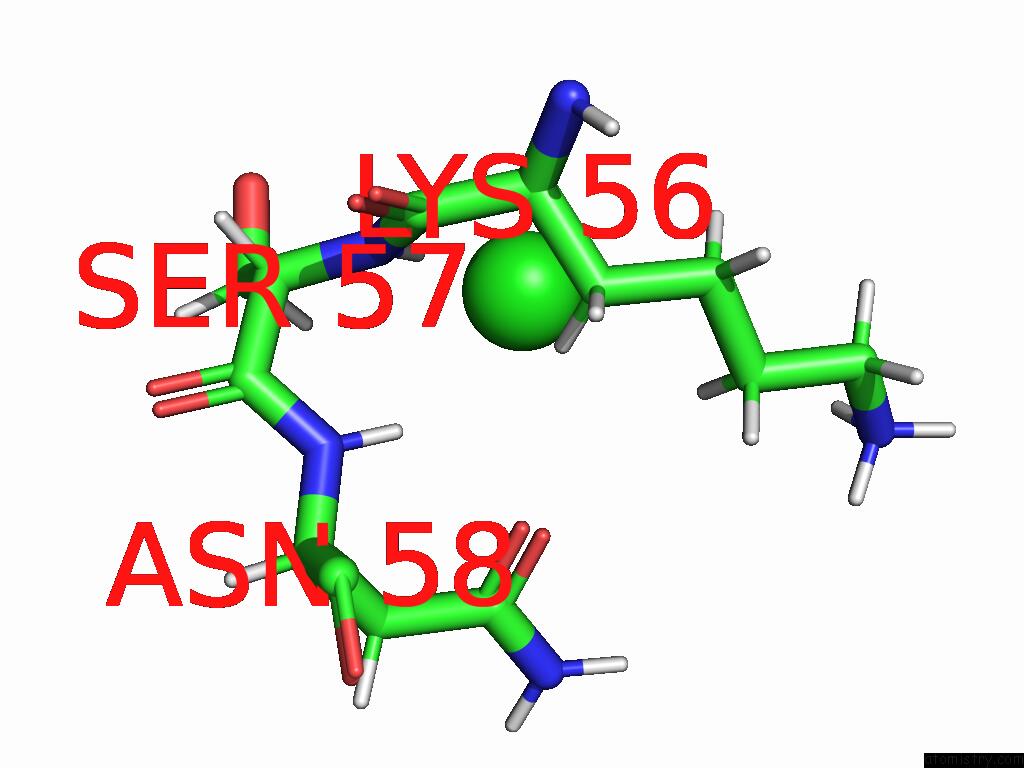
Mono view
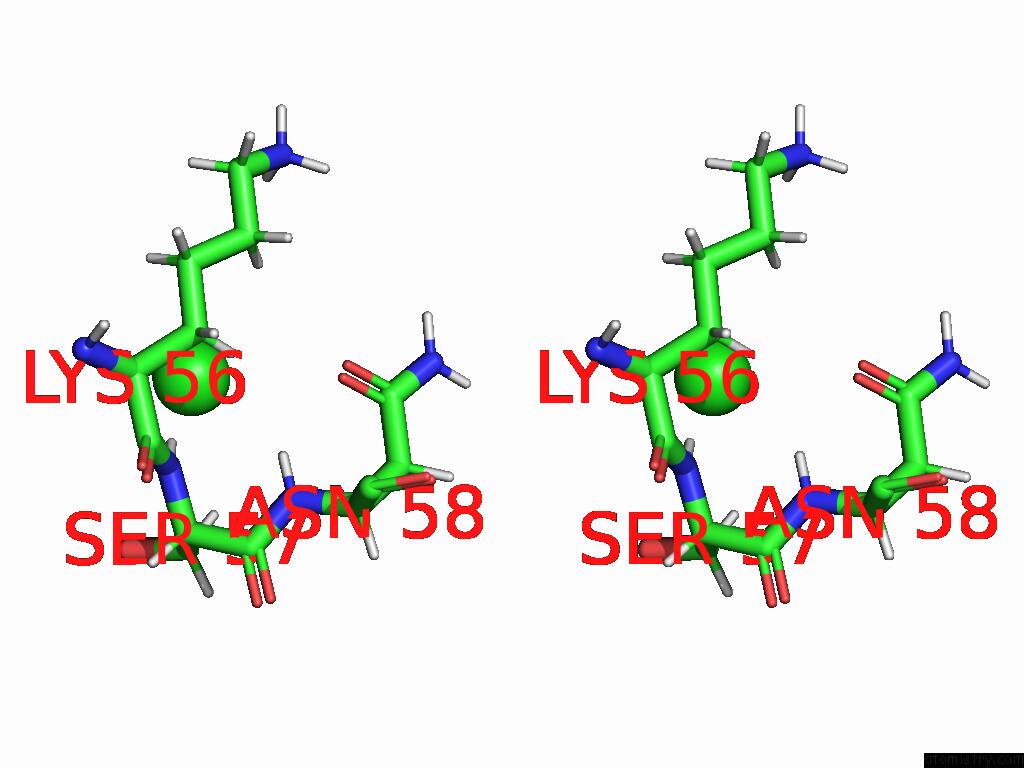
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Zea Mays Adenosine Kinase 3 (ZMADK3) in Complex with AP5A within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 8rpa
Go back to
Chlorine binding site 2 out
of 2 in the Crystal Structure of Zea Mays Adenosine Kinase 3 (ZMADK3) in Complex with AP5A
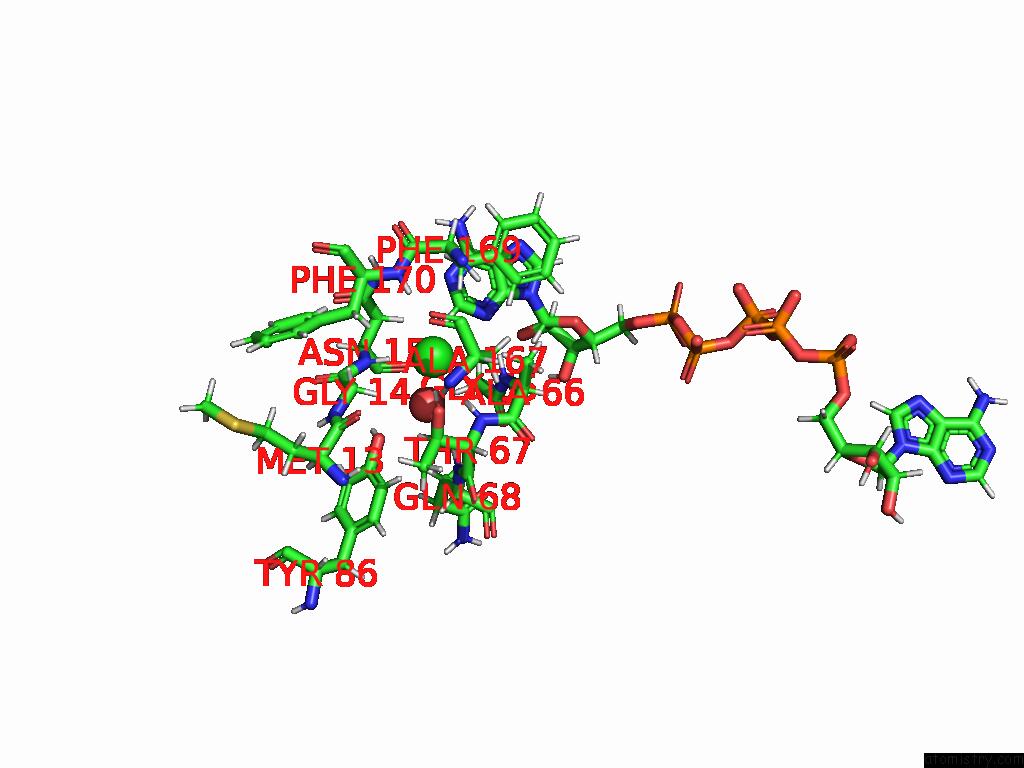
Mono view
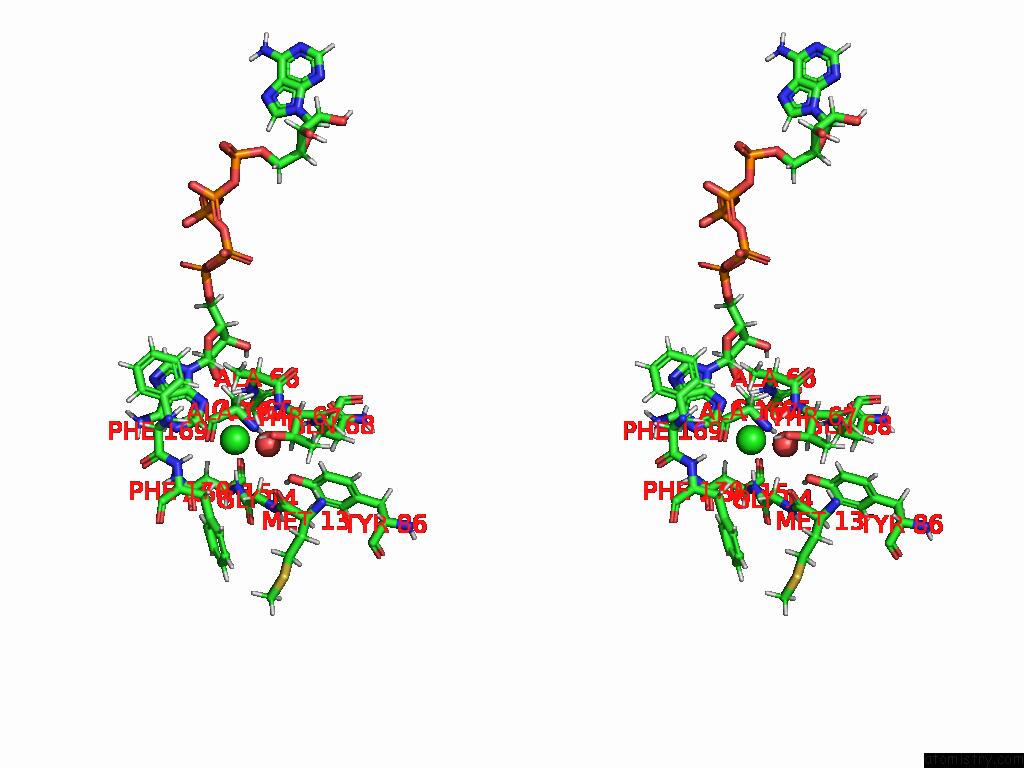
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of Zea Mays Adenosine Kinase 3 (ZMADK3) in Complex with AP5A within 5.0Å range:
|
Reference:
D.J.Kopecny,
A.Vigouroux,
J.Belicek,
M.Kopecna,
R.Koncitikova,
I.Petrik,
V.Mik,
J.F.Humplik,
M.Le Berre,
S.Plancqueel,
M.Strnad,
K.Von Schwartzenberg,
O.Novak,
S.Morera,
D.Kopecny.
Structure-Function Study on Plant Adenosine Kinase Phosphorylating Adenosine and Cytokinin Ribosides To Be Published.
Page generated: Sun Jul 13 13:58:41 2025
Last articles
K in 6NLGK in 6NC1
K in 6NFV
K in 6NFU
K in 6NAX
K in 6N95
K in 6N94
K in 6N93
K in 6MZ2
K in 6N65