Chlorine »
PDB 8rx0-8sd4 »
8s1p »
Chlorine in PDB 8s1p: Ylmh Bound to Ptrna-50S
Other elements in 8s1p:
The structure of Ylmh Bound to Ptrna-50S also contains other interesting chemical elements:
| Zinc | (Zn) | 4 atoms |
| Potassium | (K) | 47 atoms |
| Magnesium | (Mg) | 158 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Ylmh Bound to Ptrna-50S
(pdb code 8s1p). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Ylmh Bound to Ptrna-50S, PDB code: 8s1p:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Ylmh Bound to Ptrna-50S, PDB code: 8s1p:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 8s1p
Go back to
Chlorine binding site 1 out
of 2 in the Ylmh Bound to Ptrna-50S
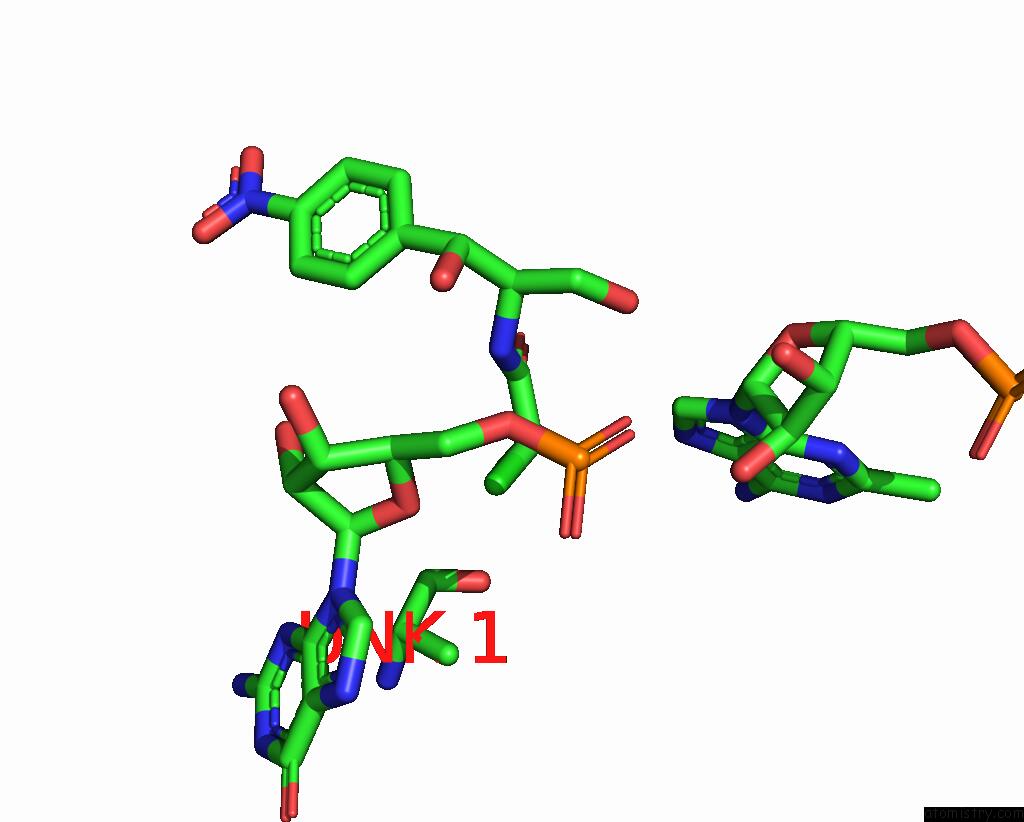
Mono view
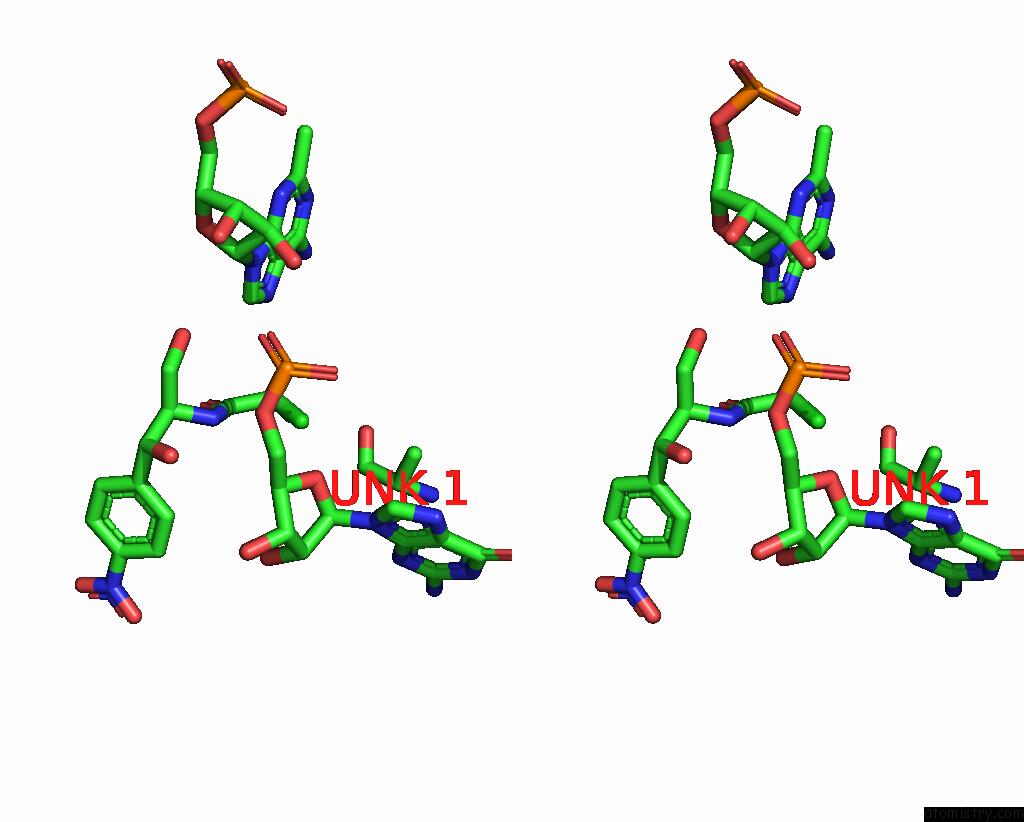
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Ylmh Bound to Ptrna-50S within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 8s1p
Go back to
Chlorine binding site 2 out
of 2 in the Ylmh Bound to Ptrna-50S
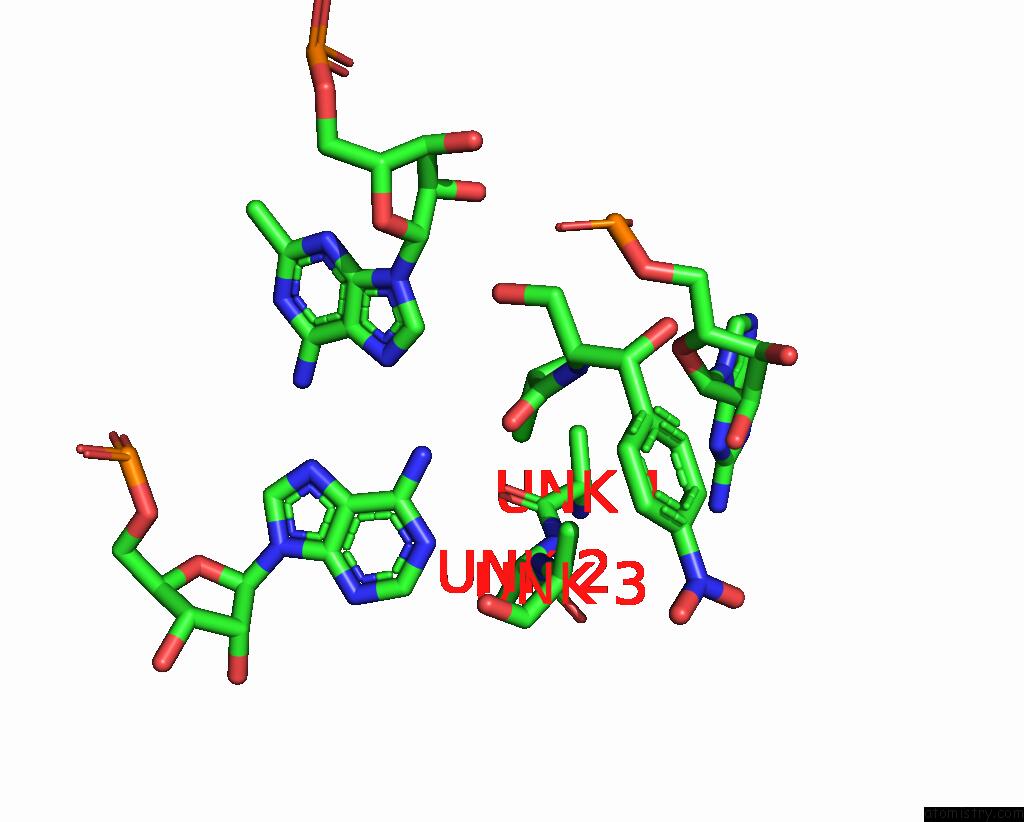
Mono view
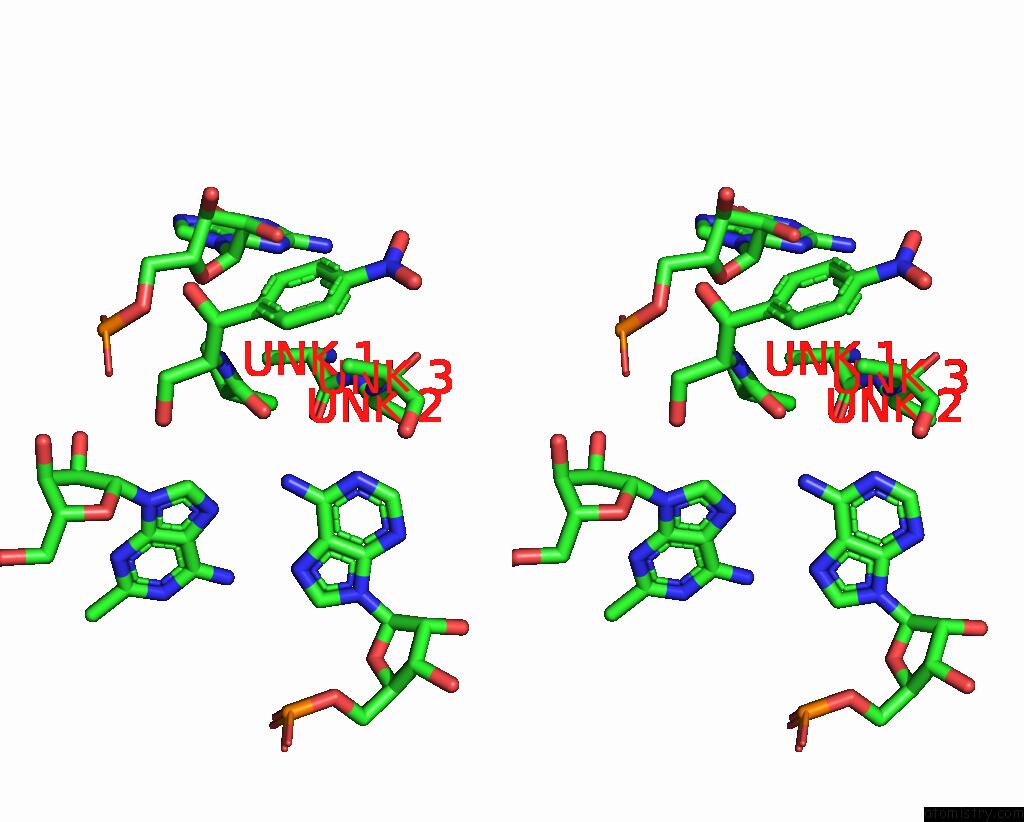
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Ylmh Bound to Ptrna-50S within 5.0Å range:
|
Reference:
H.Takada,
H.Paternoga,
K.Fujiwara,
J.A.Nakamoto,
E.N.Park,
L.Dimitrova-Paternoga,
B.Beckert,
M.Saarma,
T.Tenson,
A.R.Buskirk,
G.C.Atkinson,
S.Chiba,
D.N.Wilson,
V.Hauryliuk.
A Role For the S4-Domain Containing Protein Ylmh in Ribosome-Associated Quality Control in Bacillus Subtilis. Nucleic Acids Res. 2024.
ISSN: ESSN 1362-4962
PubMed: 38811035
DOI: 10.1093/NAR/GKAE399
Page generated: Sun Jul 13 14:02:53 2025
ISSN: ESSN 1362-4962
PubMed: 38811035
DOI: 10.1093/NAR/GKAE399
Last articles
Mg in 3KRWMg in 3KRO
Mg in 3KRF
Mg in 3KRA
Mg in 3KQL
Mg in 3KLF
Mg in 3KQI
Mg in 3KLE
Mg in 3KQ8
Mg in 3KOA