Chlorine »
PDB 8u6d-8udp »
8ucz »
Chlorine in PDB 8ucz: Sterile Alpha Motif (Sam) Domain From TRIC1 From Arabidopsis Thaliana - D235A Mutant
Protein crystallography data
The structure of Sterile Alpha Motif (Sam) Domain From TRIC1 From Arabidopsis Thaliana - D235A Mutant, PDB code: 8ucz
was solved by
B.Olasz,
A.Vrielink,
L.Smithers,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 47.05 / 2.07 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 28.676, 92.587, 94.109, 90, 90, 90 |
| R / Rfree (%) | 24.8 / 27.7 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Sterile Alpha Motif (Sam) Domain From TRIC1 From Arabidopsis Thaliana - D235A Mutant
(pdb code 8ucz). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 3 binding sites of Chlorine where determined in the Sterile Alpha Motif (Sam) Domain From TRIC1 From Arabidopsis Thaliana - D235A Mutant, PDB code: 8ucz:
Jump to Chlorine binding site number: 1; 2; 3;
In total 3 binding sites of Chlorine where determined in the Sterile Alpha Motif (Sam) Domain From TRIC1 From Arabidopsis Thaliana - D235A Mutant, PDB code: 8ucz:
Jump to Chlorine binding site number: 1; 2; 3;
Chlorine binding site 1 out of 3 in 8ucz
Go back to
Chlorine binding site 1 out
of 3 in the Sterile Alpha Motif (Sam) Domain From TRIC1 From Arabidopsis Thaliana - D235A Mutant
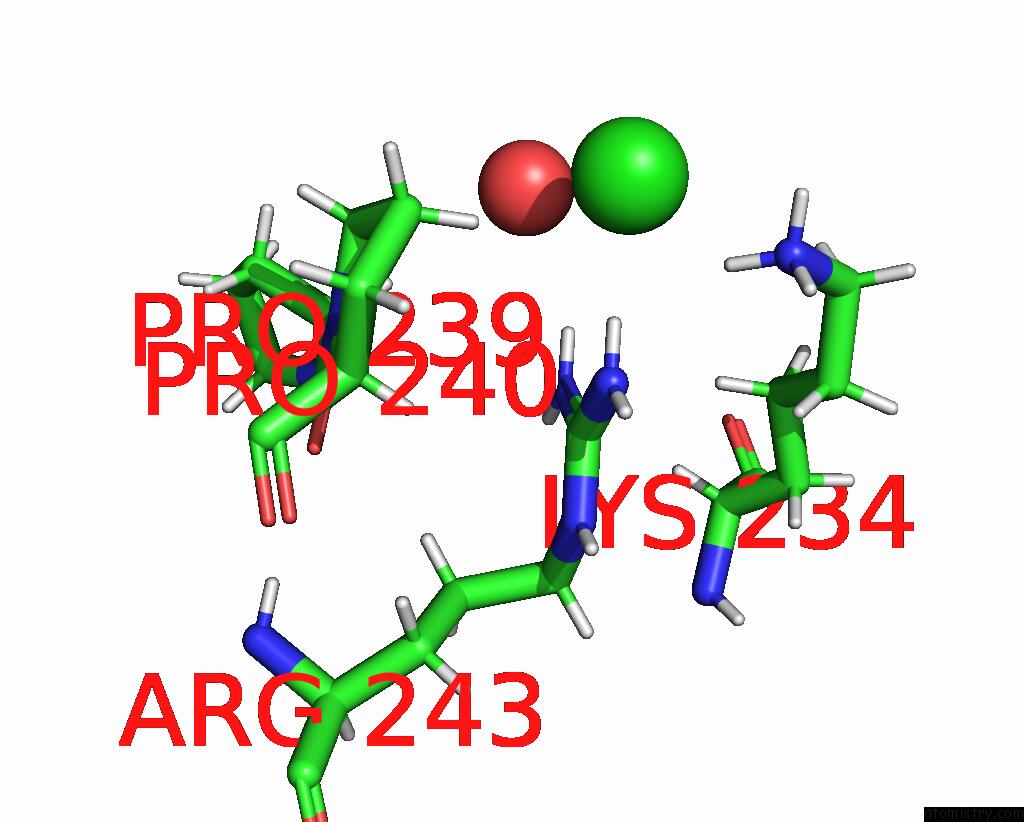
Mono view
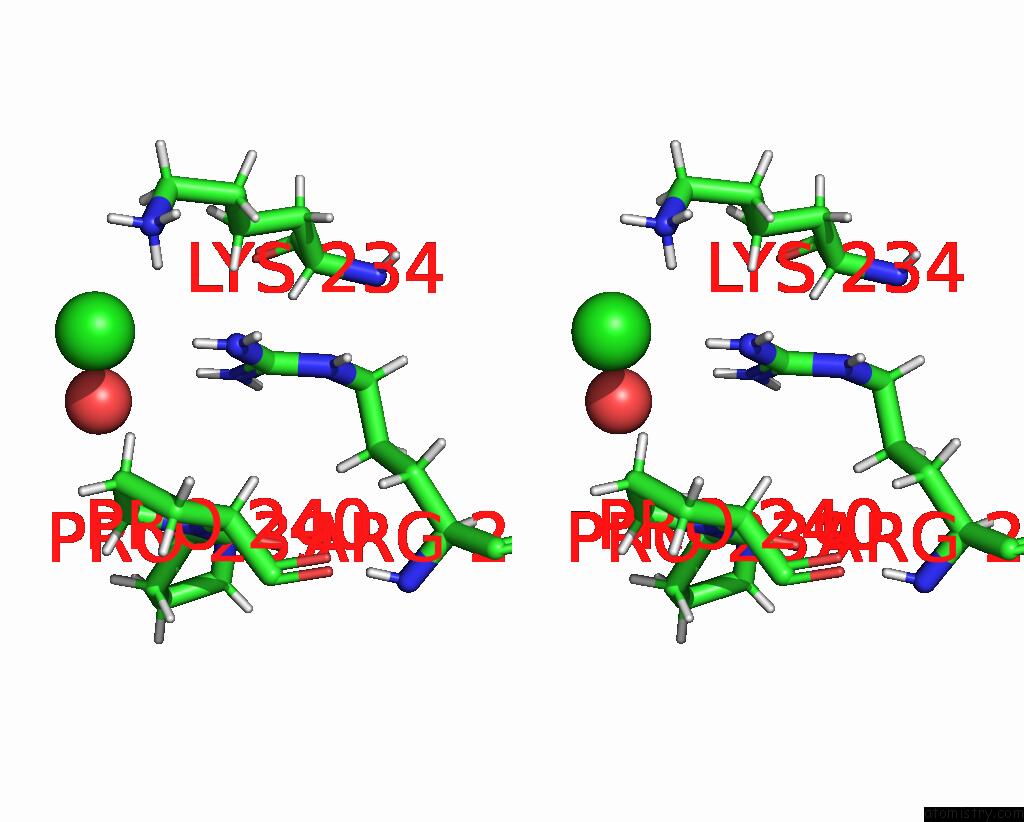
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Sterile Alpha Motif (Sam) Domain From TRIC1 From Arabidopsis Thaliana - D235A Mutant within 5.0Å range:
|
Chlorine binding site 2 out of 3 in 8ucz
Go back to
Chlorine binding site 2 out
of 3 in the Sterile Alpha Motif (Sam) Domain From TRIC1 From Arabidopsis Thaliana - D235A Mutant
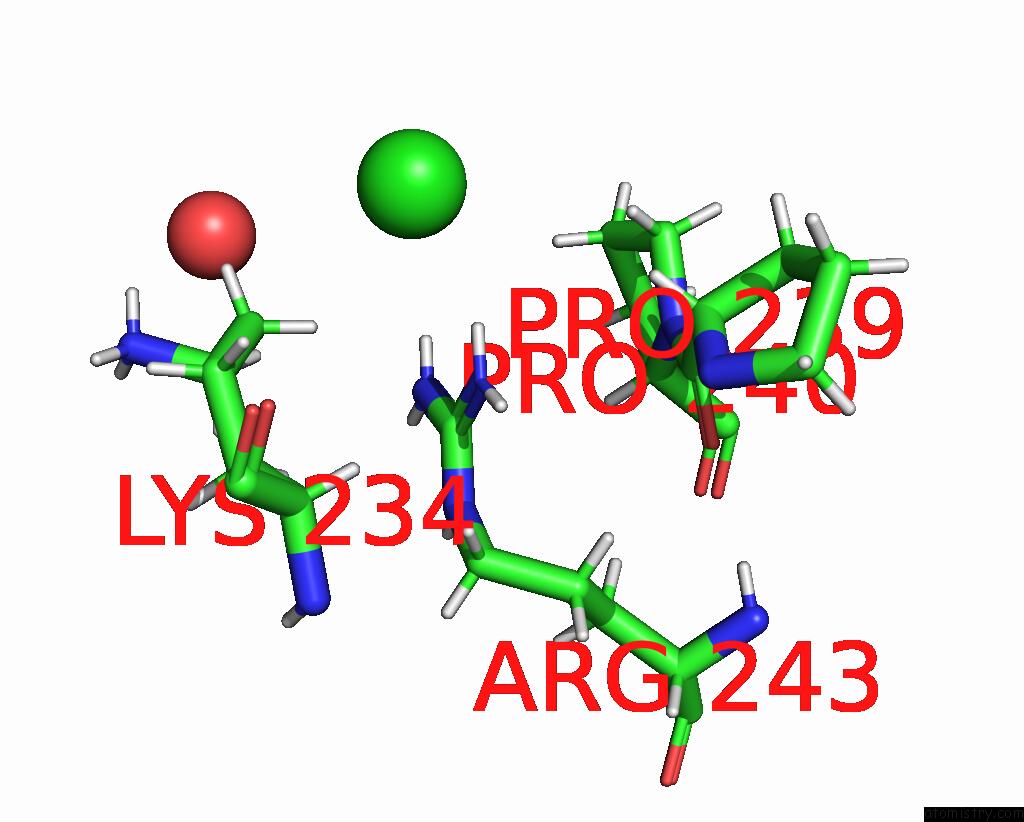
Mono view
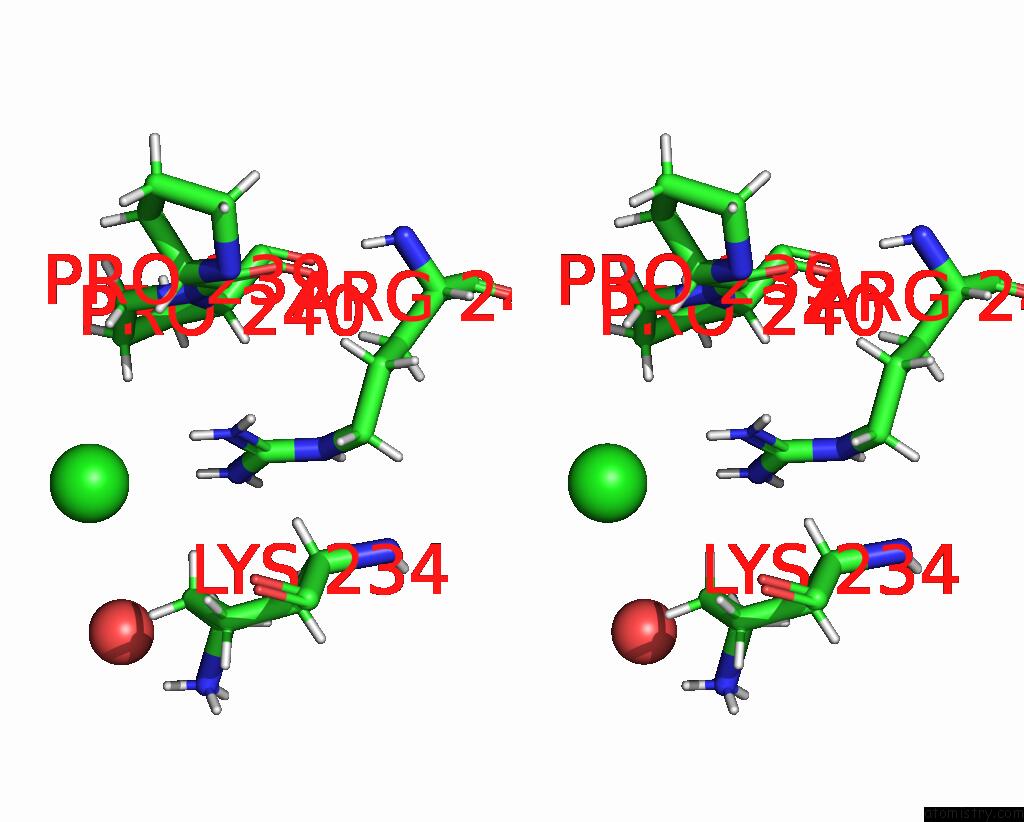
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Sterile Alpha Motif (Sam) Domain From TRIC1 From Arabidopsis Thaliana - D235A Mutant within 5.0Å range:
|
Chlorine binding site 3 out of 3 in 8ucz
Go back to
Chlorine binding site 3 out
of 3 in the Sterile Alpha Motif (Sam) Domain From TRIC1 From Arabidopsis Thaliana - D235A Mutant
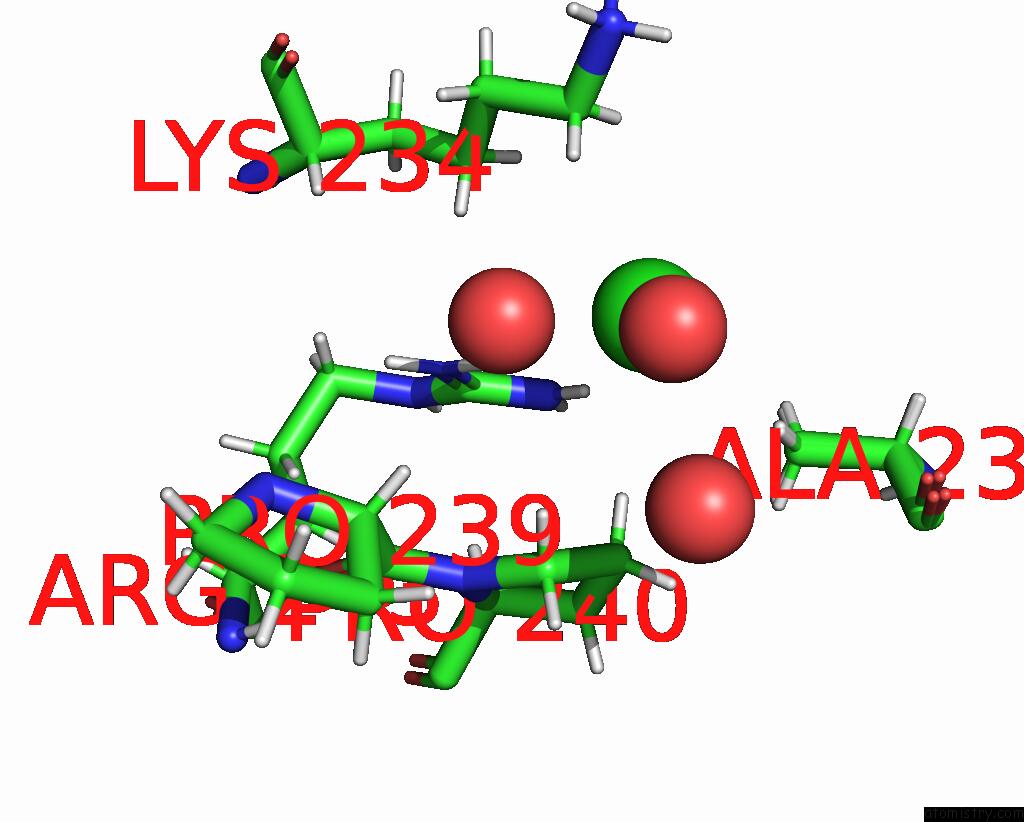
Mono view
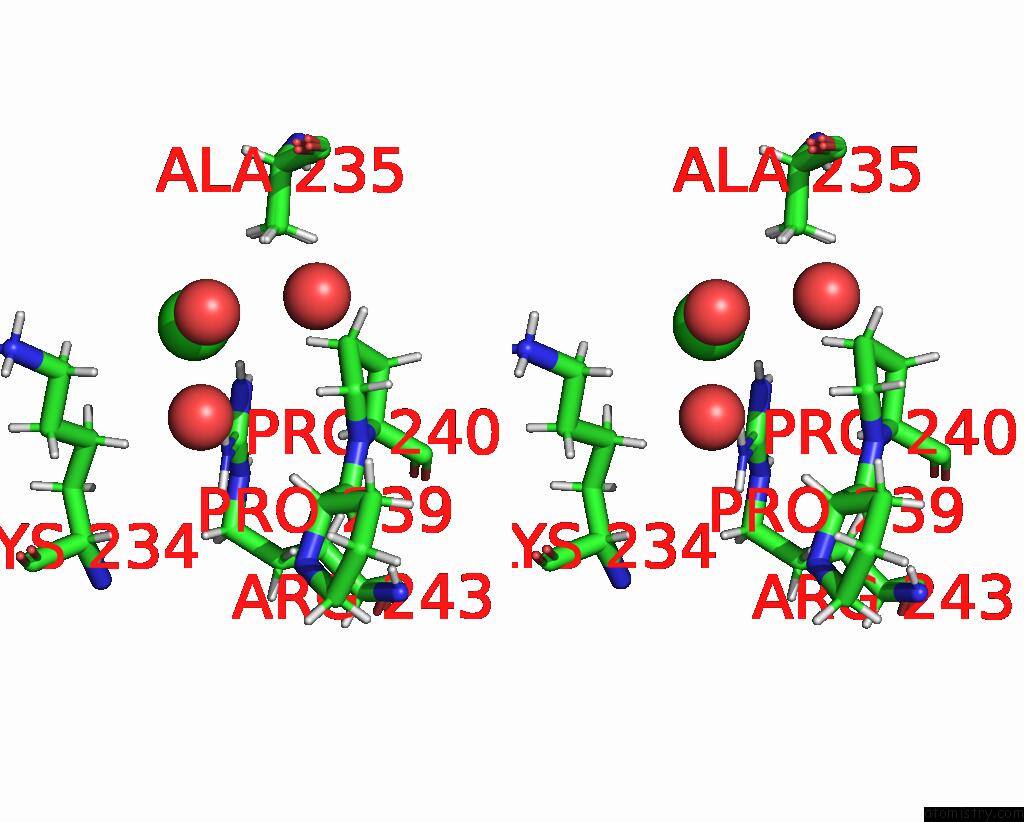
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Sterile Alpha Motif (Sam) Domain From TRIC1 From Arabidopsis Thaliana - D235A Mutant within 5.0Å range:
|
Reference:
B.Olasz,
L.Smithers,
G.L.Evans,
A.Anandan,
M.W.Murcha,
A.Vrielink.
Structural Analysis of the Sam Domain of the Arabidopsis Mitochondrial Trna Import Receptor. J.Biol.Chem. 07258 2024.
ISSN: ESSN 1083-351X
PubMed: 38582448
DOI: 10.1016/J.JBC.2024.107258
Page generated: Sun Jul 13 14:50:34 2025
ISSN: ESSN 1083-351X
PubMed: 38582448
DOI: 10.1016/J.JBC.2024.107258
Last articles
K in 5EDUK in 5EEK
K in 5EC2
K in 5EE9
K in 5DKP
K in 5EC1
K in 5EBW
K in 5EBL
K in 5EBM
K in 5DOU