Chlorine »
PDB 8wby-8wtr »
8wcr »
Chlorine in PDB 8wcr: Cryo-Em Structure of Nanodisc (Pe:Ps:Pc) Reconstituted Glic at pH 4 in Open State
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Cryo-Em Structure of Nanodisc (Pe:Ps:Pc) Reconstituted Glic at pH 4 in Open State
(pdb code 8wcr). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 5 binding sites of Chlorine where determined in the Cryo-Em Structure of Nanodisc (Pe:Ps:Pc) Reconstituted Glic at pH 4 in Open State, PDB code: 8wcr:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Chlorine where determined in the Cryo-Em Structure of Nanodisc (Pe:Ps:Pc) Reconstituted Glic at pH 4 in Open State, PDB code: 8wcr:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5;
Chlorine binding site 1 out of 5 in 8wcr
Go back to
Chlorine binding site 1 out
of 5 in the Cryo-Em Structure of Nanodisc (Pe:Ps:Pc) Reconstituted Glic at pH 4 in Open State
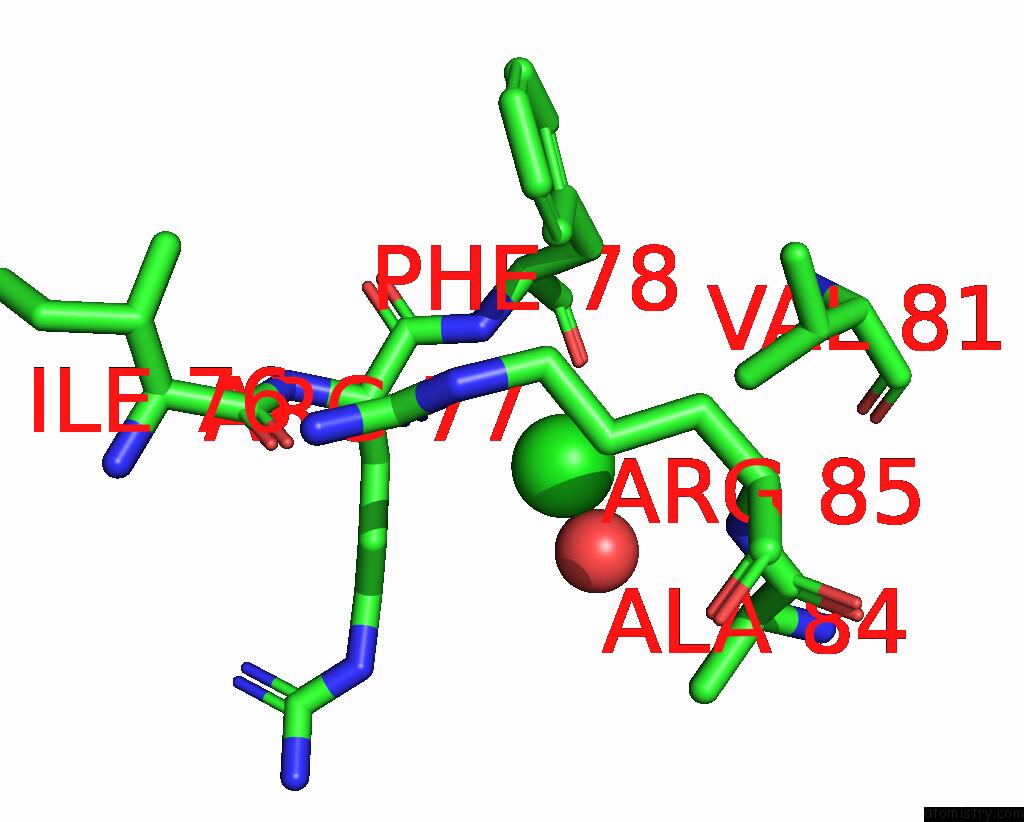
Mono view
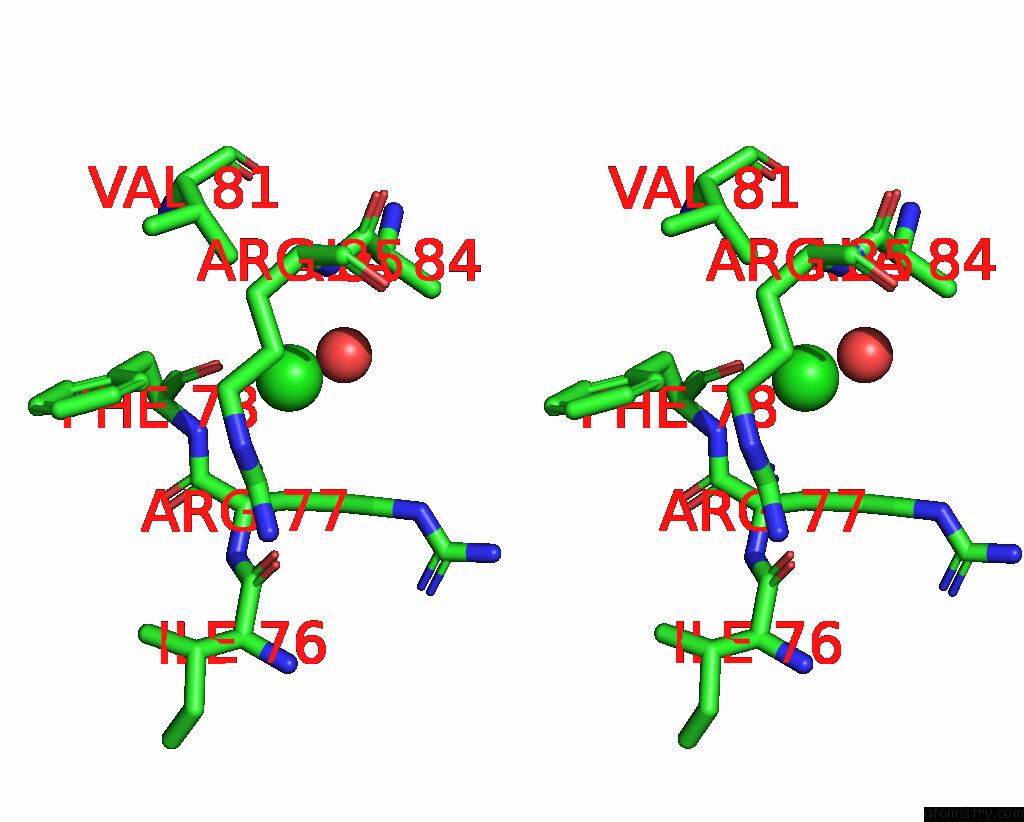
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Cryo-Em Structure of Nanodisc (Pe:Ps:Pc) Reconstituted Glic at pH 4 in Open State within 5.0Å range:
|
Chlorine binding site 2 out of 5 in 8wcr
Go back to
Chlorine binding site 2 out
of 5 in the Cryo-Em Structure of Nanodisc (Pe:Ps:Pc) Reconstituted Glic at pH 4 in Open State
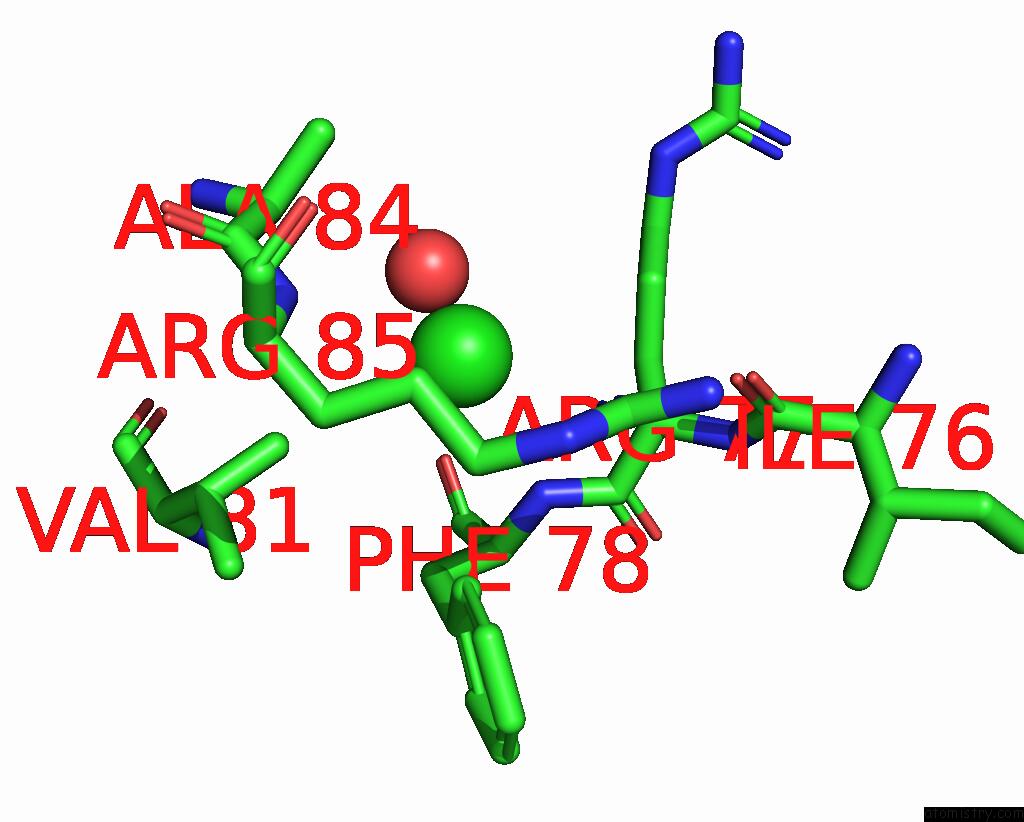
Mono view
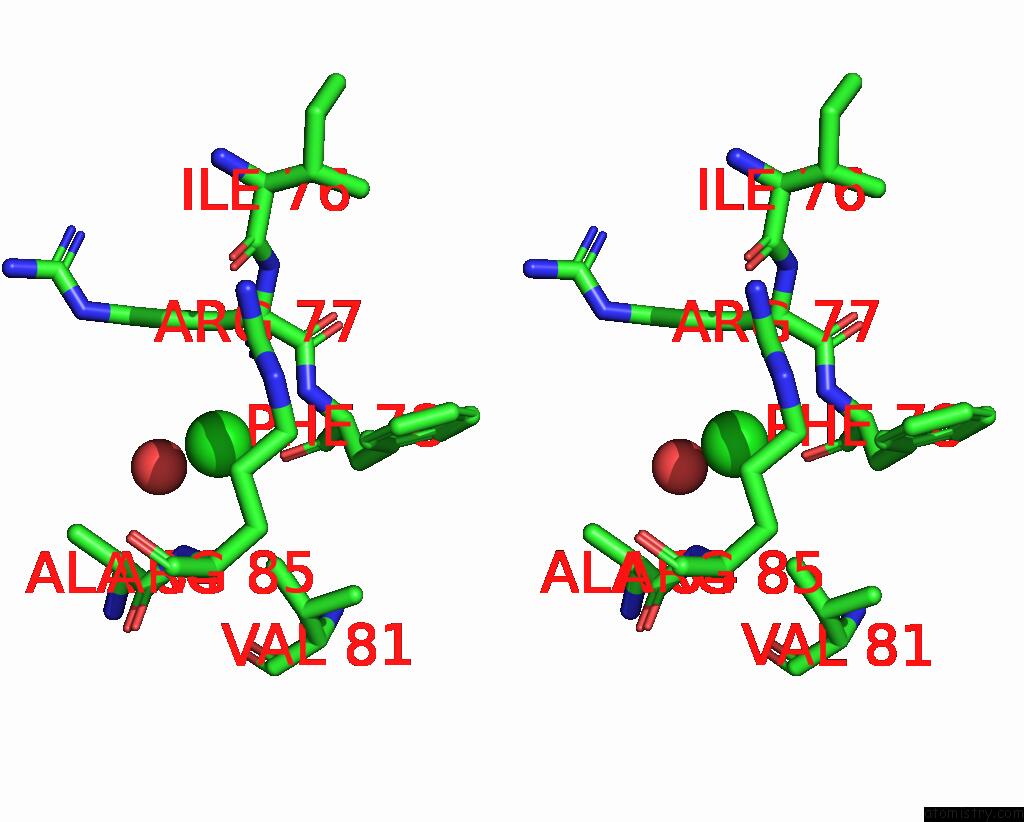
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Cryo-Em Structure of Nanodisc (Pe:Ps:Pc) Reconstituted Glic at pH 4 in Open State within 5.0Å range:
|
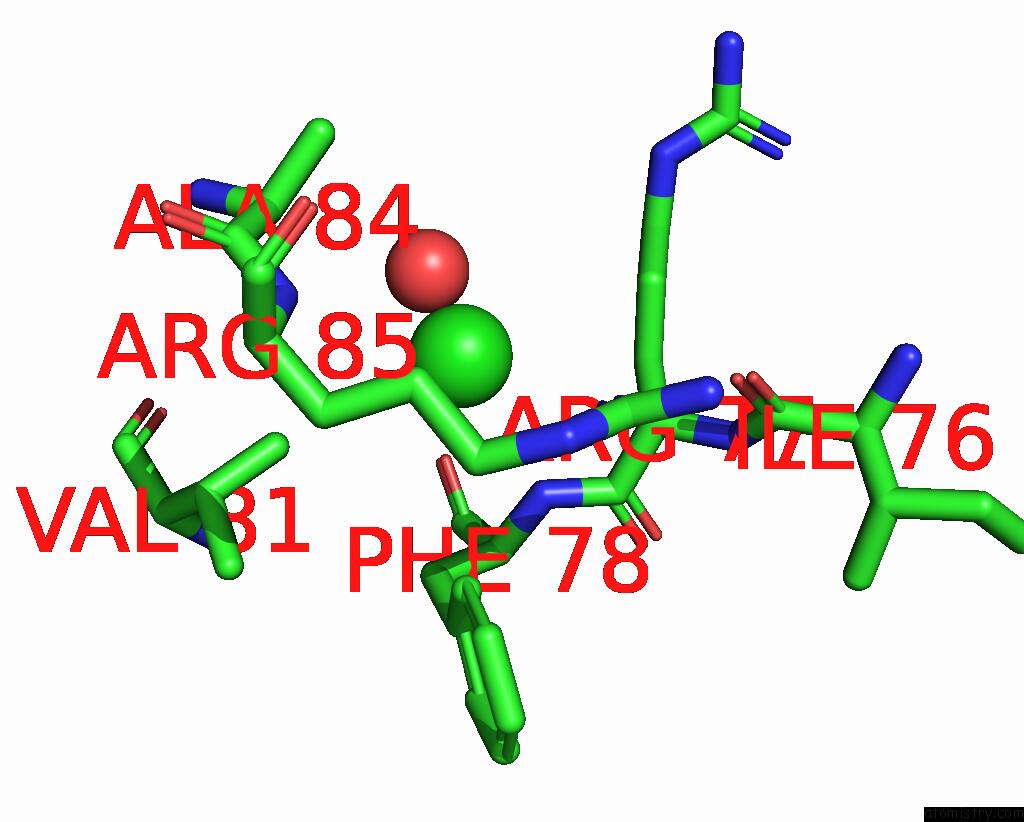
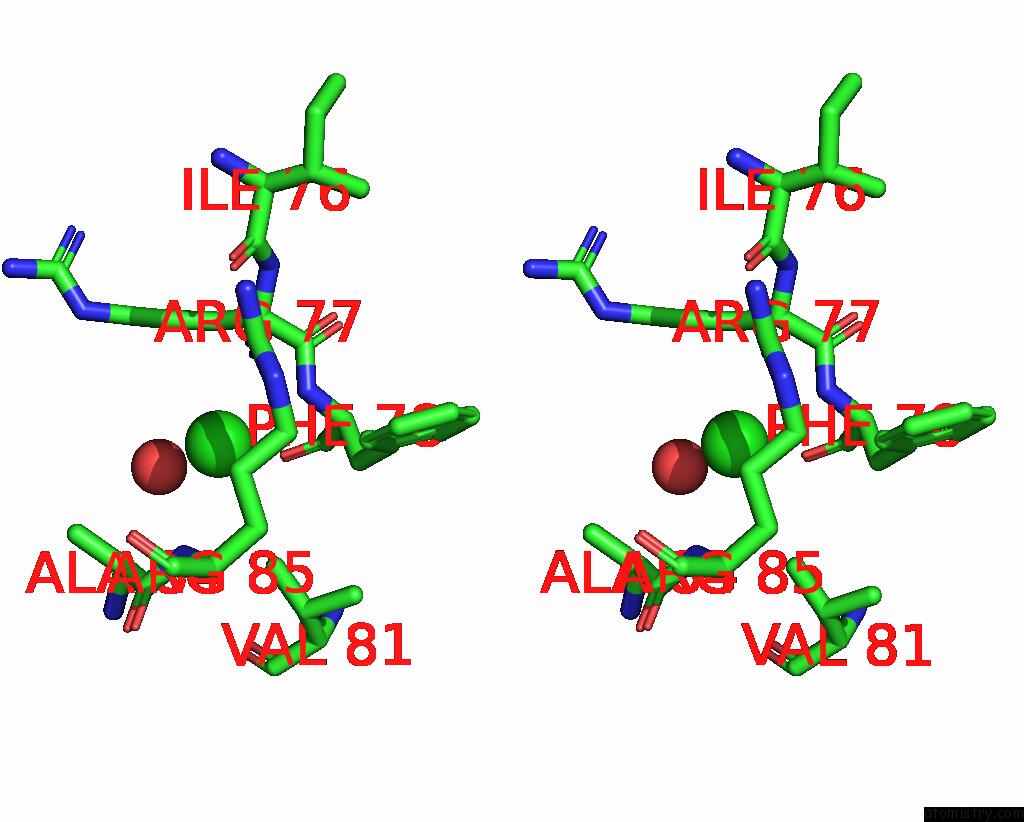
Chlorine binding site 3 out of 5 in 8wcr
Go back to
Chlorine binding site 3 out
of 5 in the Cryo-Em Structure of Nanodisc (Pe:Ps:Pc) Reconstituted Glic at pH 4 in Open State
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Cryo-Em Structure of Nanodisc (Pe:Ps:Pc) Reconstituted Glic at pH 4 in Open State within 5.0Å range:
|
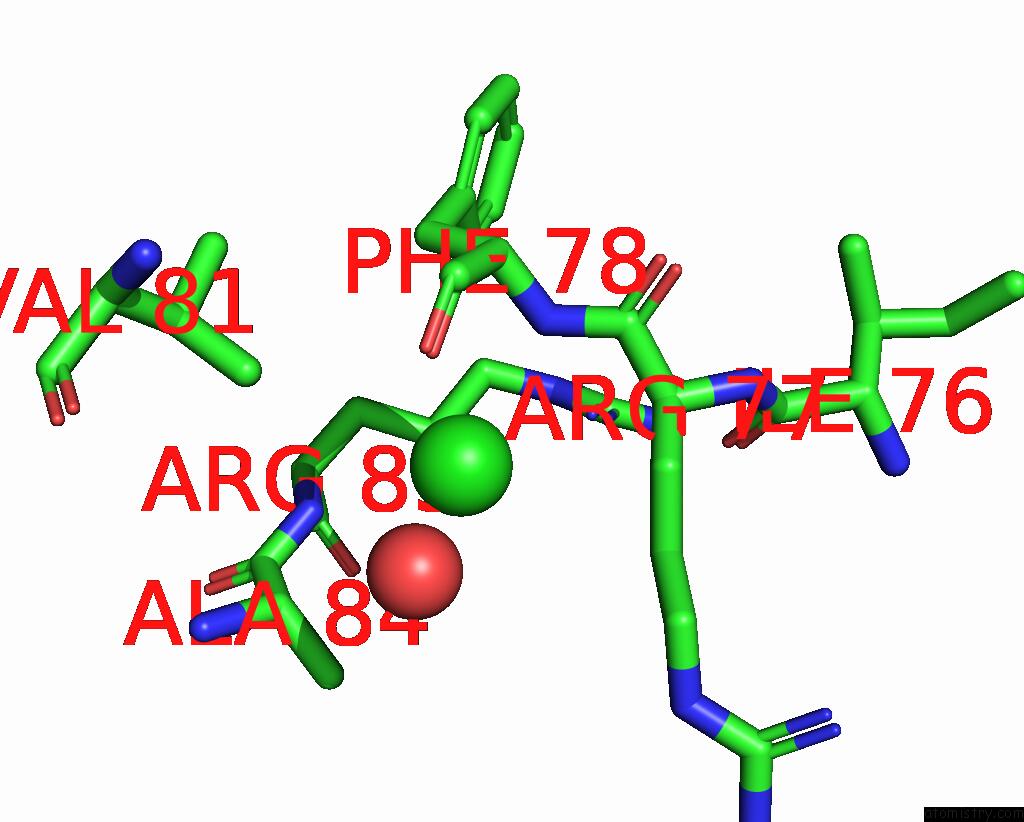
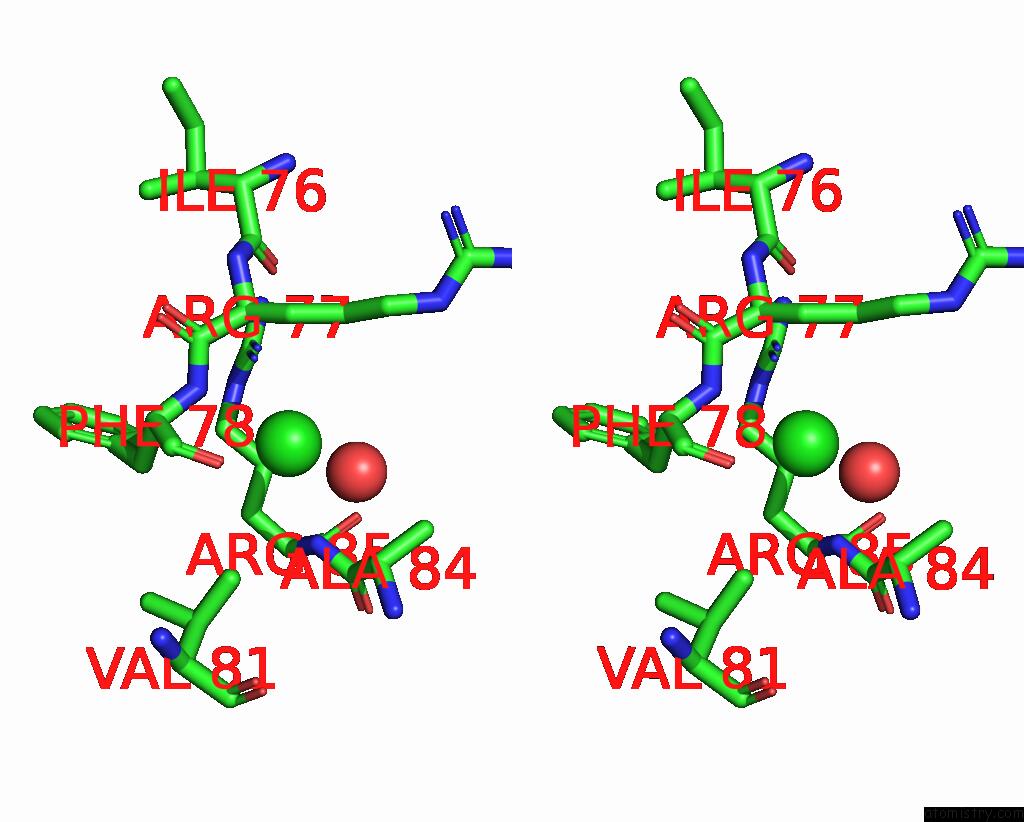
Chlorine binding site 4 out of 5 in 8wcr
Go back to
Chlorine binding site 4 out
of 5 in the Cryo-Em Structure of Nanodisc (Pe:Ps:Pc) Reconstituted Glic at pH 4 in Open State
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Cryo-Em Structure of Nanodisc (Pe:Ps:Pc) Reconstituted Glic at pH 4 in Open State within 5.0Å range:
|
Chlorine binding site 5 out of 5 in 8wcr
Go back to
Chlorine binding site 5 out
of 5 in the Cryo-Em Structure of Nanodisc (Pe:Ps:Pc) Reconstituted Glic at pH 4 in Open State
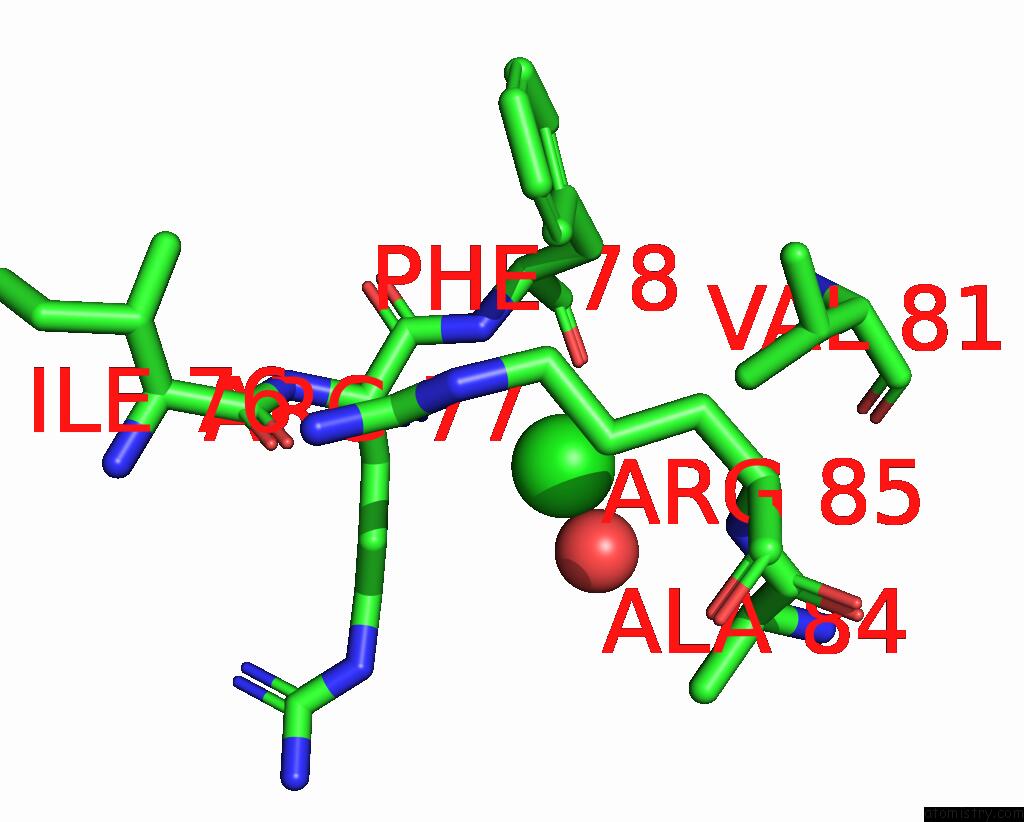
Mono view
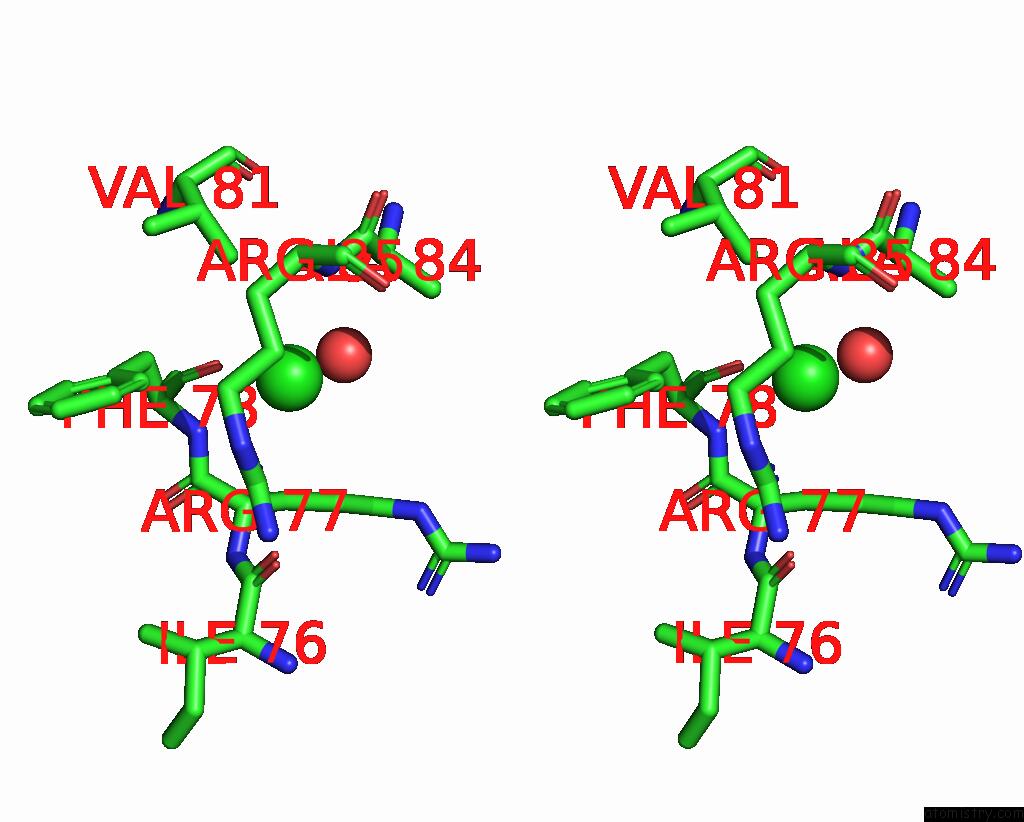
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Cryo-Em Structure of Nanodisc (Pe:Ps:Pc) Reconstituted Glic at pH 4 in Open State within 5.0Å range:
|
Reference:
N.Bharambe,
Z.Li,
D.Seiferth,
A.M.Balakrishna,
P.C.Biggin,
S.Basak.
Cryo-Em Structures of Prokaryotic Ligand-Gated Ion Channel Glic Provide Insights Into Gating in A Lipid Environment. Nat Commun V. 15 2967 2024.
ISSN: ESSN 2041-1723
PubMed: 38580666
DOI: 10.1038/S41467-024-47370-W
Page generated: Sun Jul 13 15:32:08 2025
ISSN: ESSN 2041-1723
PubMed: 38580666
DOI: 10.1038/S41467-024-47370-W
Last articles
K in 2QYOK in 2RKB
K in 2R9R
K in 2SGE
K in 2SGD
K in 2RKY
K in 2RDG
K in 2RGQ
K in 2QKS
K in 2QXL