Chlorine »
PDB 8xet-8yq4 »
8yak »
Chlorine in PDB 8yak: Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase
Other elements in 8yak:
The structure of Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase also contains other interesting chemical elements:
| Zinc | (Zn) | 16 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase
(pdb code 8yak). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 8 binding sites of Chlorine where determined in the Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase, PDB code: 8yak:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
In total 8 binding sites of Chlorine where determined in the Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase, PDB code: 8yak:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
Chlorine binding site 1 out of 8 in 8yak
Go back to
Chlorine binding site 1 out
of 8 in the Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase
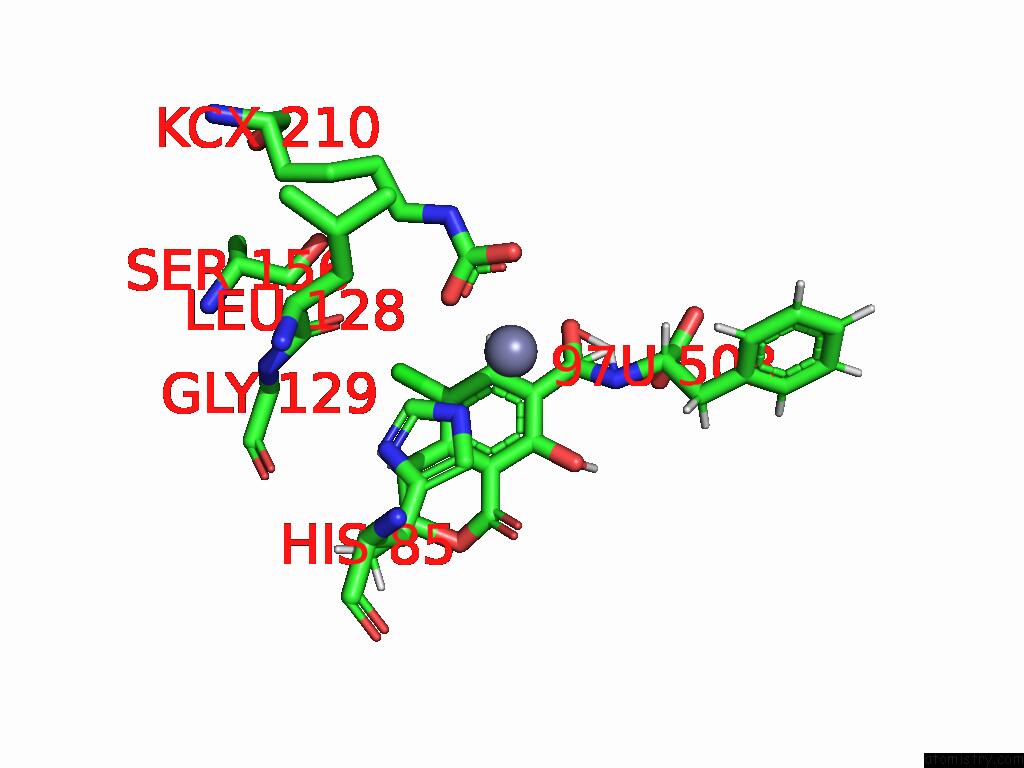
Mono view
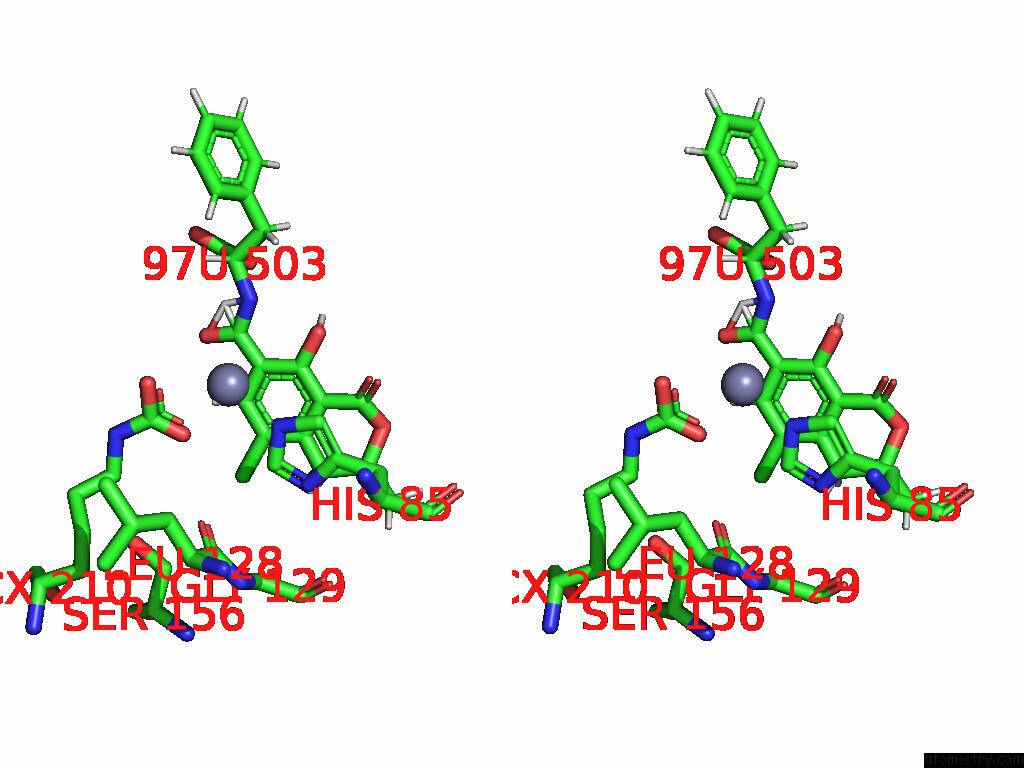
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase within 5.0Å range:
|
Chlorine binding site 2 out of 8 in 8yak
Go back to
Chlorine binding site 2 out
of 8 in the Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase
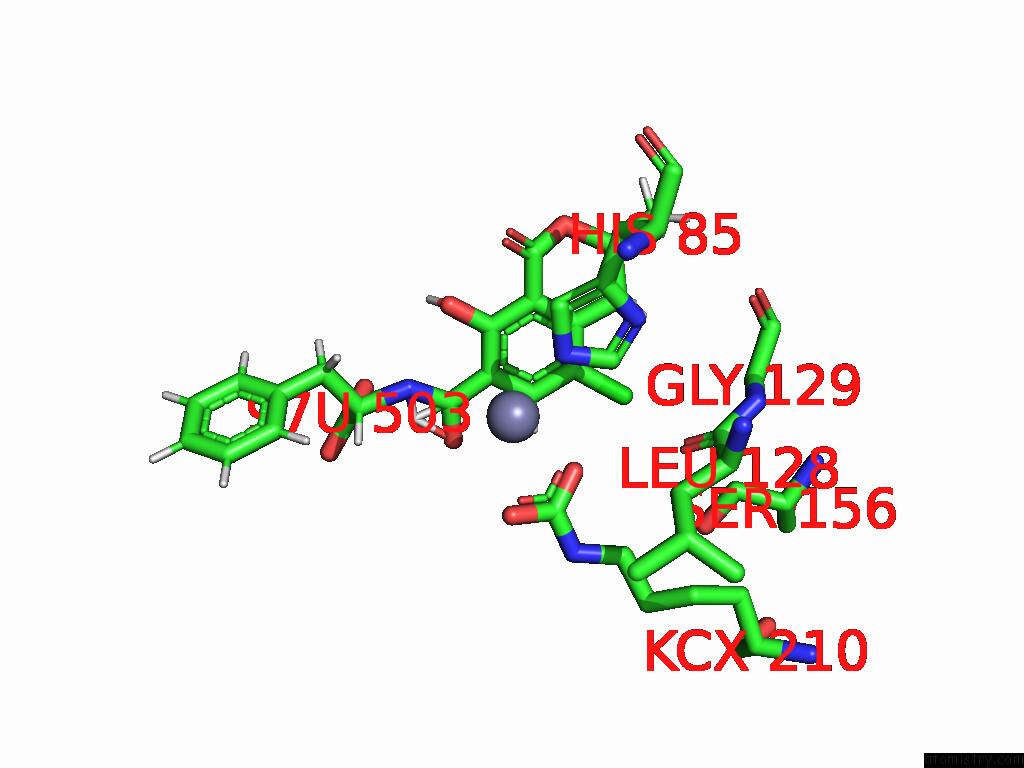
Mono view
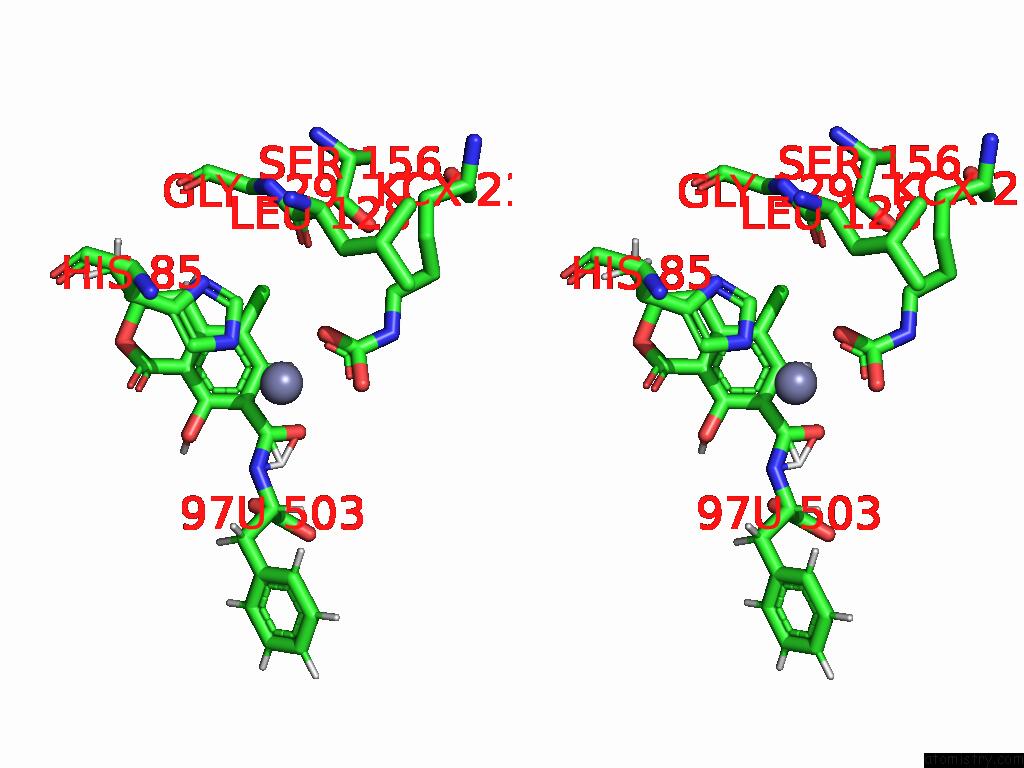
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase within 5.0Å range:
|
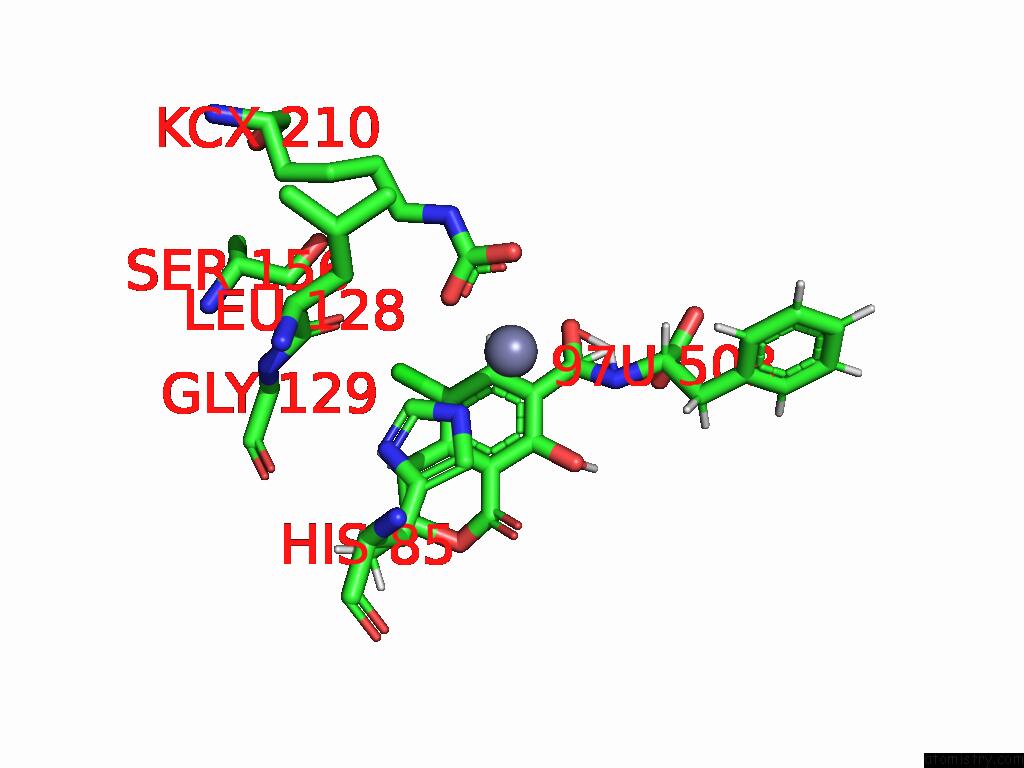
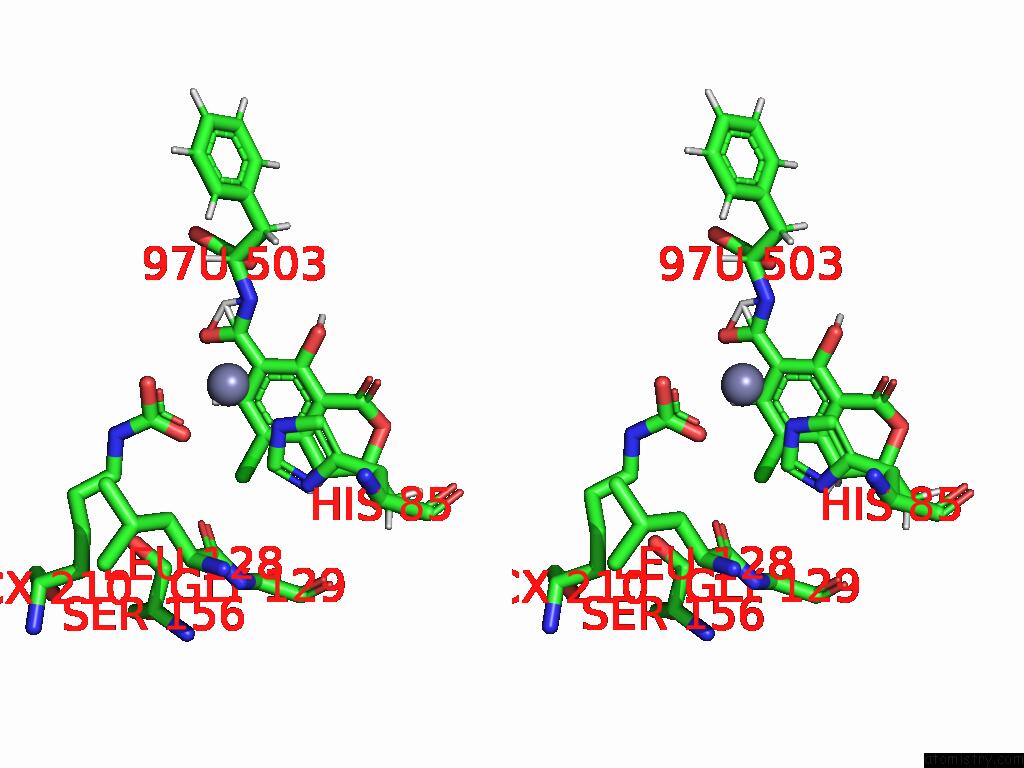
Chlorine binding site 3 out of 8 in 8yak
Go back to
Chlorine binding site 3 out
of 8 in the Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase within 5.0Å range:
|
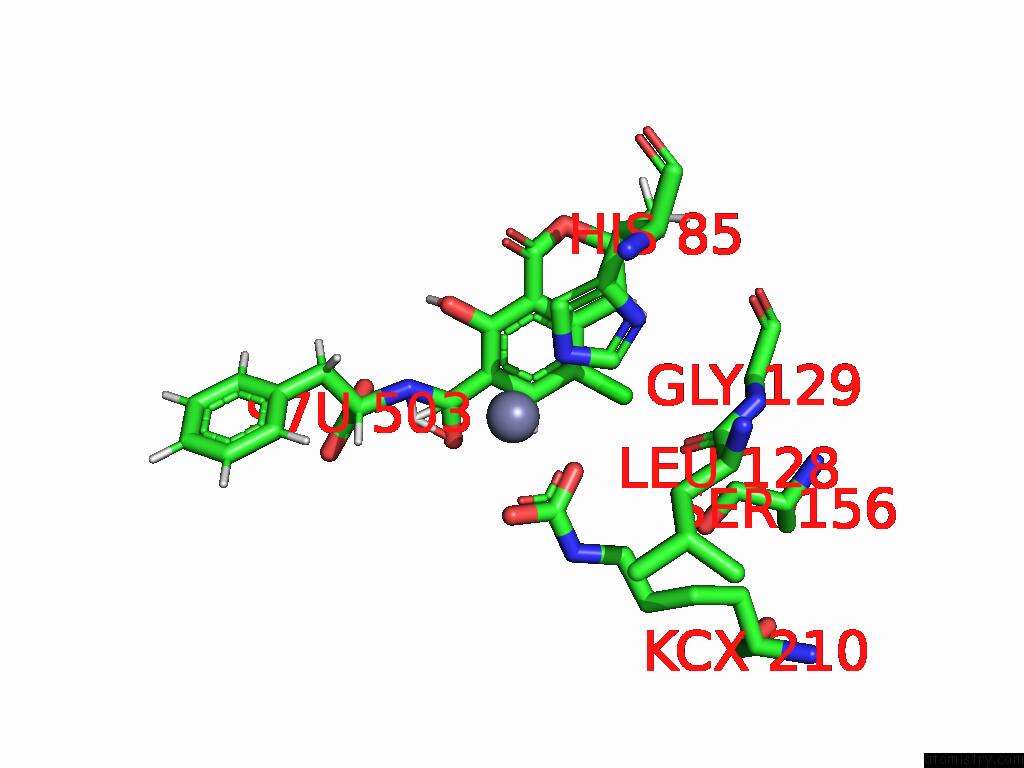
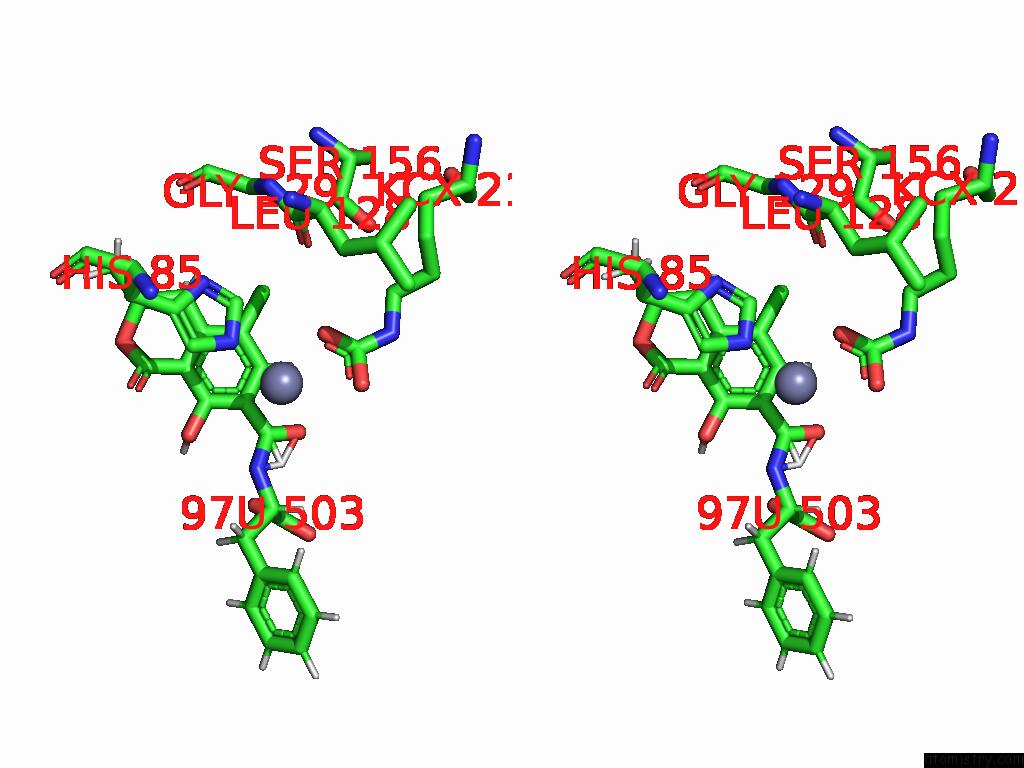
Chlorine binding site 4 out of 8 in 8yak
Go back to
Chlorine binding site 4 out
of 8 in the Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase within 5.0Å range:
|
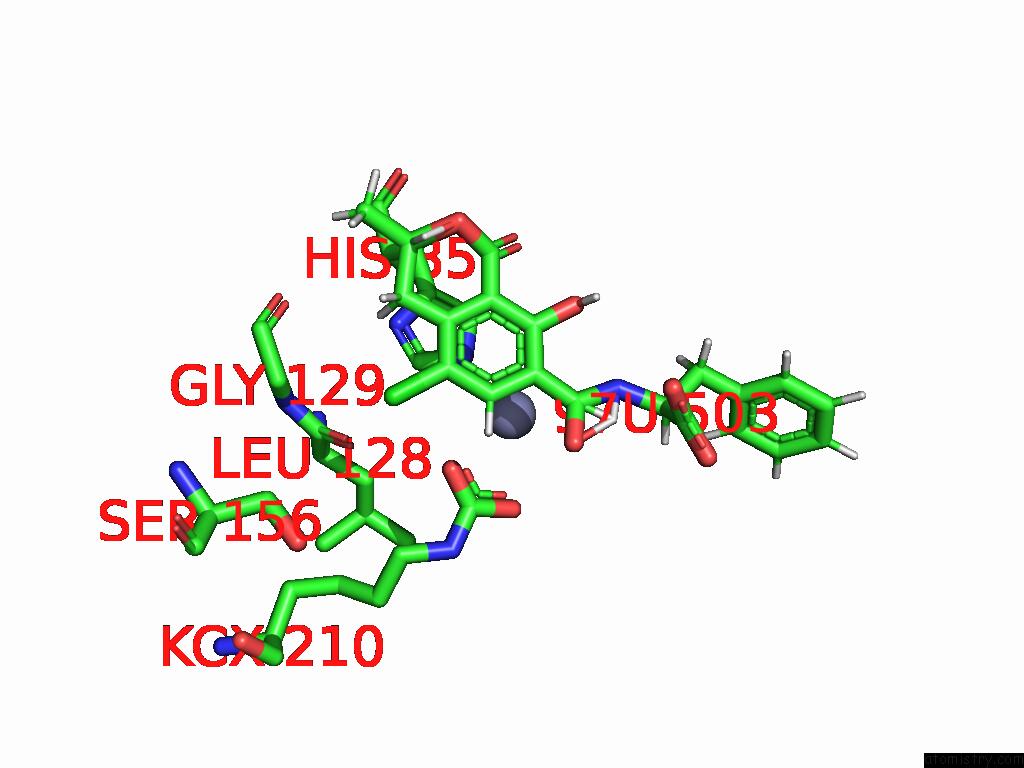
Chlorine binding site 5 out of 8 in 8yak
Go back to
Chlorine binding site 5 out
of 8 in the Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase within 5.0Å range:
|
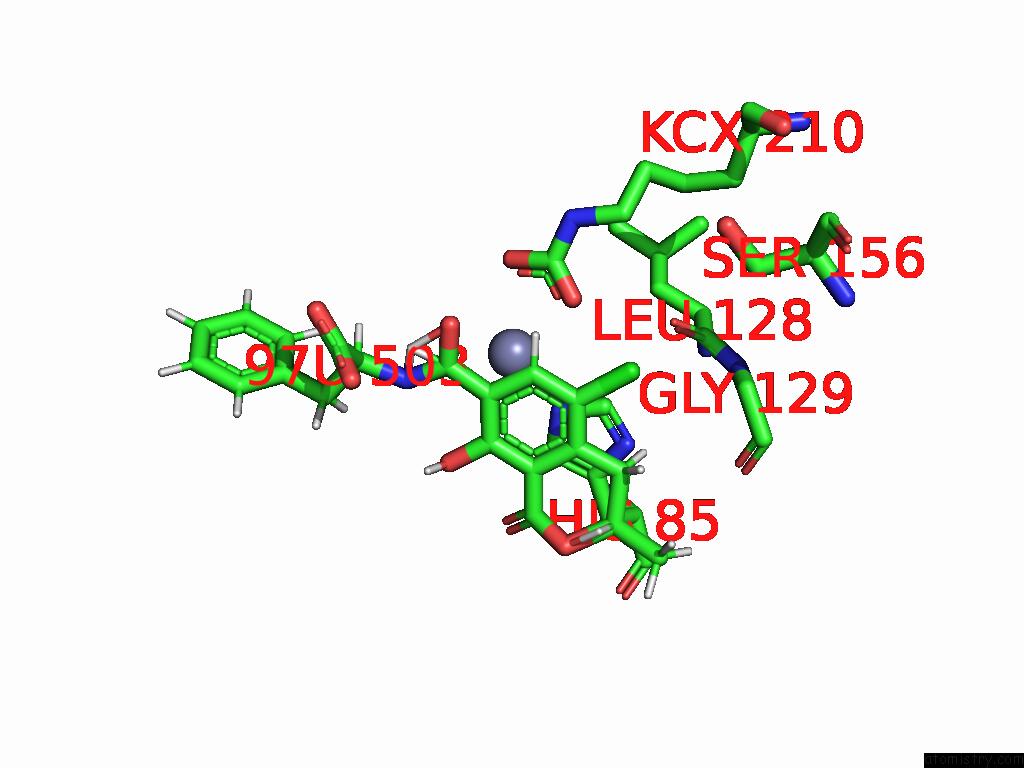
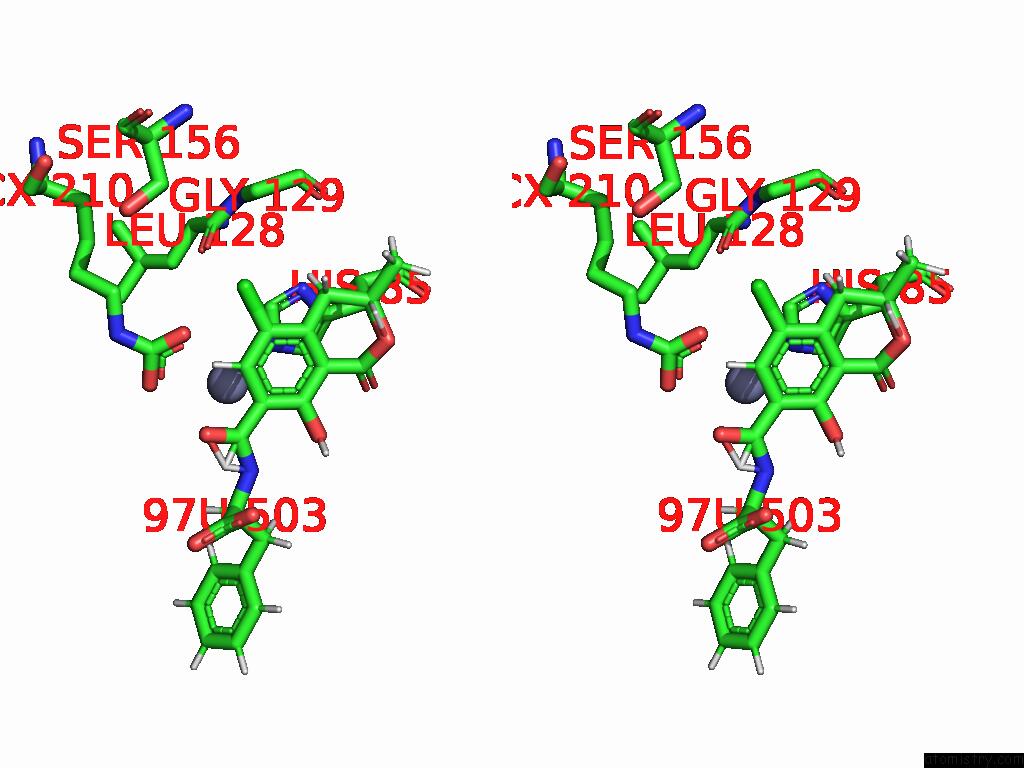
Chlorine binding site 6 out of 8 in 8yak
Go back to
Chlorine binding site 6 out
of 8 in the Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase within 5.0Å range:
|
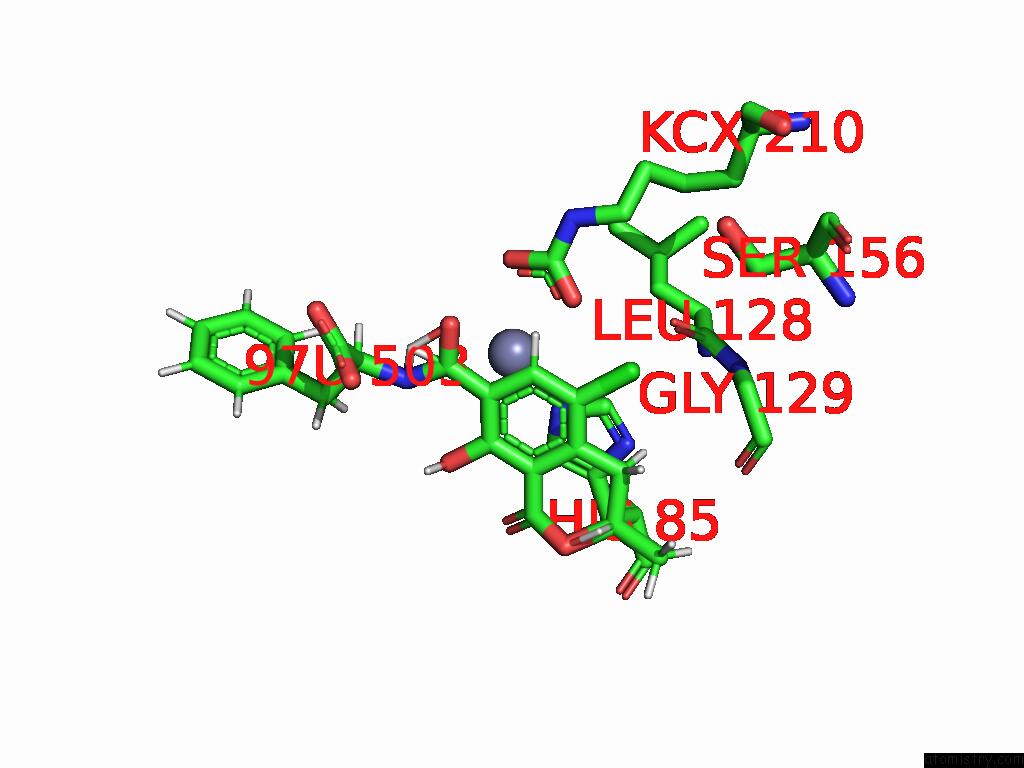
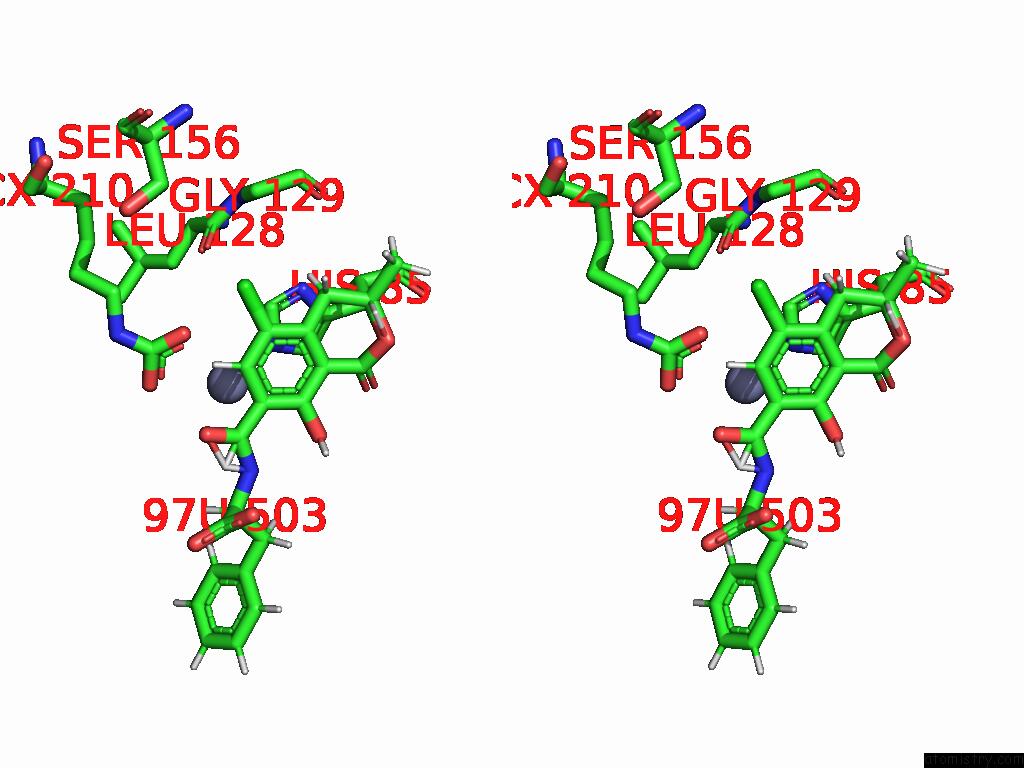
Chlorine binding site 7 out of 8 in 8yak
Go back to
Chlorine binding site 7 out
of 8 in the Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 7 of Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase within 5.0Å range:
|
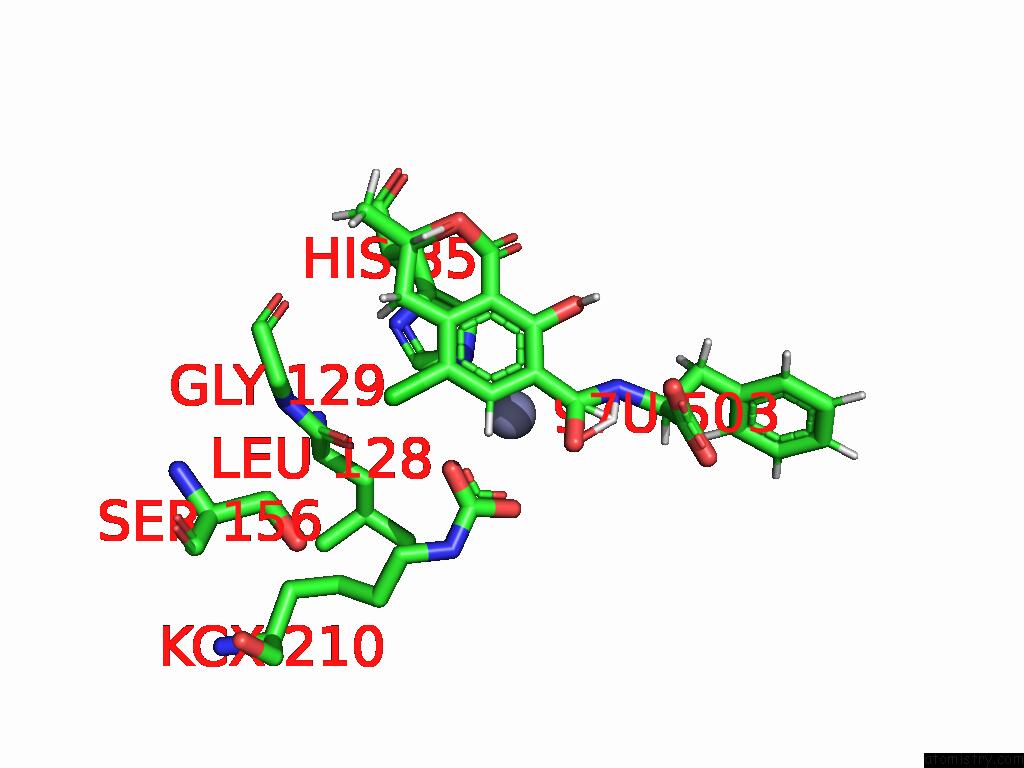
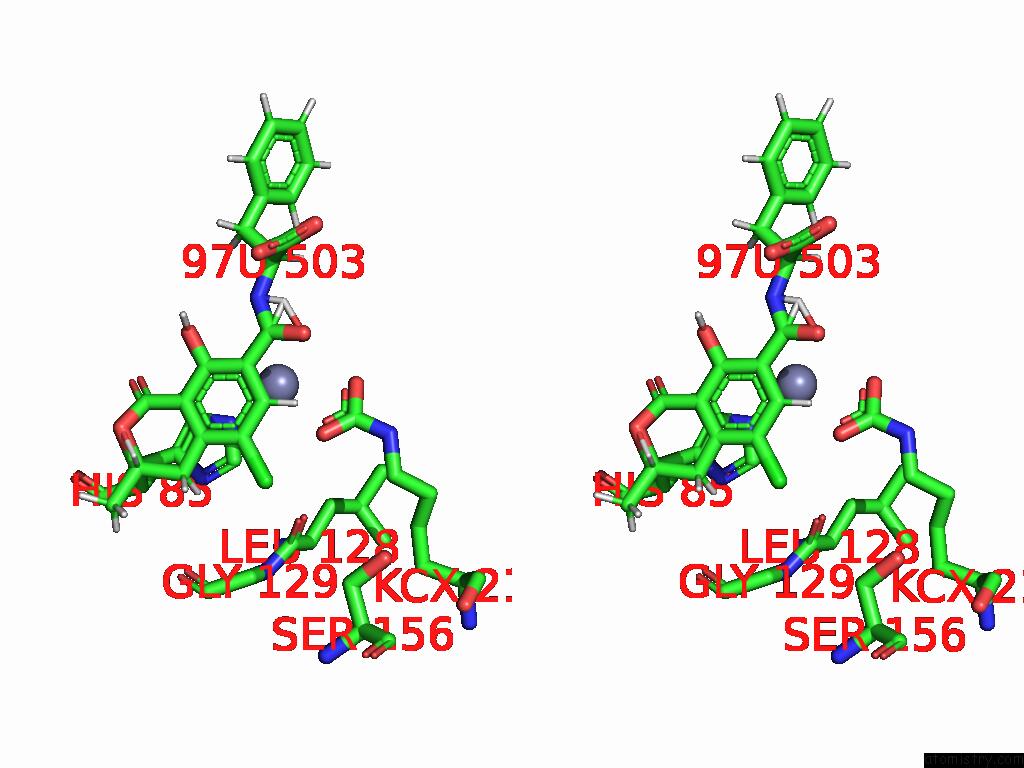
Chlorine binding site 8 out of 8 in 8yak
Go back to
Chlorine binding site 8 out
of 8 in the Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 8 of Cryo-Em Structure and Rational Engineering of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase within 5.0Å range:
|
Reference:
Y.Hu,
L.Dai,
Y.Xu,
D.Niu,
X.Yang,
Z.Xie,
P.Shen,
X.Li,
H.Li,
L.Zhang,
J.Min,
R.T.Guo,
C.C.Chen.
Functional Characterization and Structural Basis of An Efficient Ochratoxin A-Degrading Amidohydrolase. Int.J.Biol.Macromol. V. 278 34831 2024.
ISSN: ISSN 0141-8130
PubMed: 39163957
DOI: 10.1016/J.IJBIOMAC.2024.134831
Page generated: Sun Jul 13 15:45:57 2025
ISSN: ISSN 0141-8130
PubMed: 39163957
DOI: 10.1016/J.IJBIOMAC.2024.134831
Last articles
I in 6OWYI in 6OWX
I in 6OWH
I in 6OGB
I in 6OL2
I in 6O8C
I in 6O8B
I in 6O43
I in 6NYH
I in 6NQO