Chlorine »
PDB 8xet-8yq4 »
8yaq »
Chlorine in PDB 8yaq: Cryo-Em Structure of Cellodextrin Phosphorylase From Clostridium Thermocellum with Cellodextrin Ligands
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Cryo-Em Structure of Cellodextrin Phosphorylase From Clostridium Thermocellum with Cellodextrin Ligands
(pdb code 8yaq). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 5 binding sites of Chlorine where determined in the Cryo-Em Structure of Cellodextrin Phosphorylase From Clostridium Thermocellum with Cellodextrin Ligands, PDB code: 8yaq:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Chlorine where determined in the Cryo-Em Structure of Cellodextrin Phosphorylase From Clostridium Thermocellum with Cellodextrin Ligands, PDB code: 8yaq:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5;
Chlorine binding site 1 out of 5 in 8yaq
Go back to
Chlorine binding site 1 out
of 5 in the Cryo-Em Structure of Cellodextrin Phosphorylase From Clostridium Thermocellum with Cellodextrin Ligands
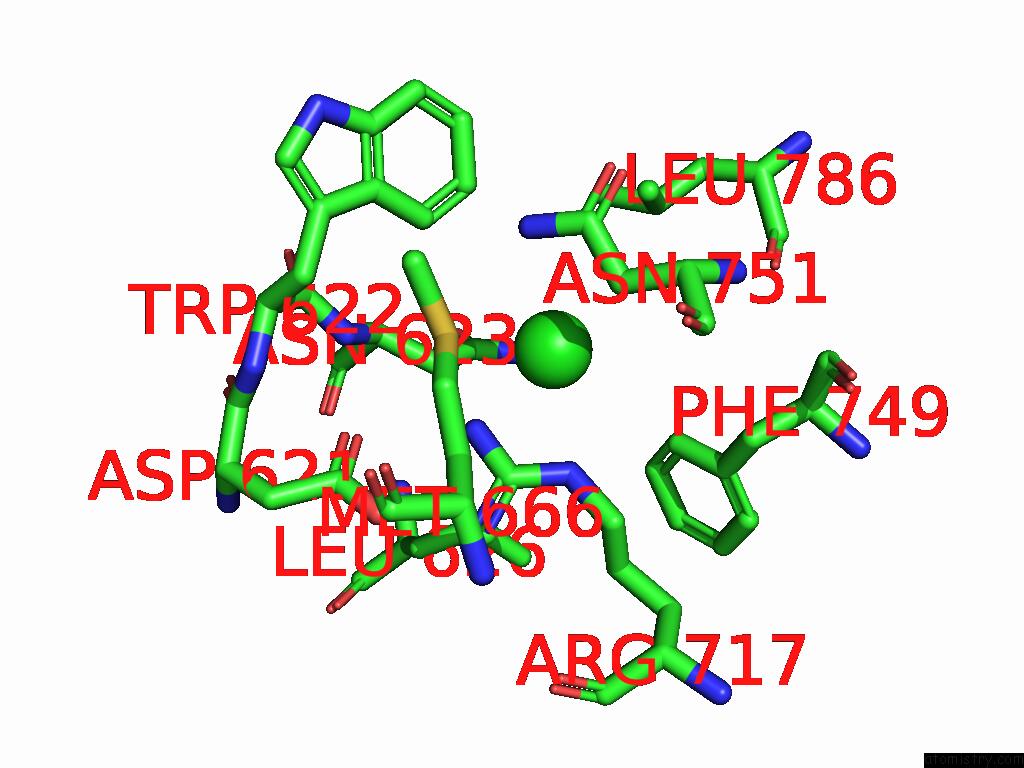
Mono view
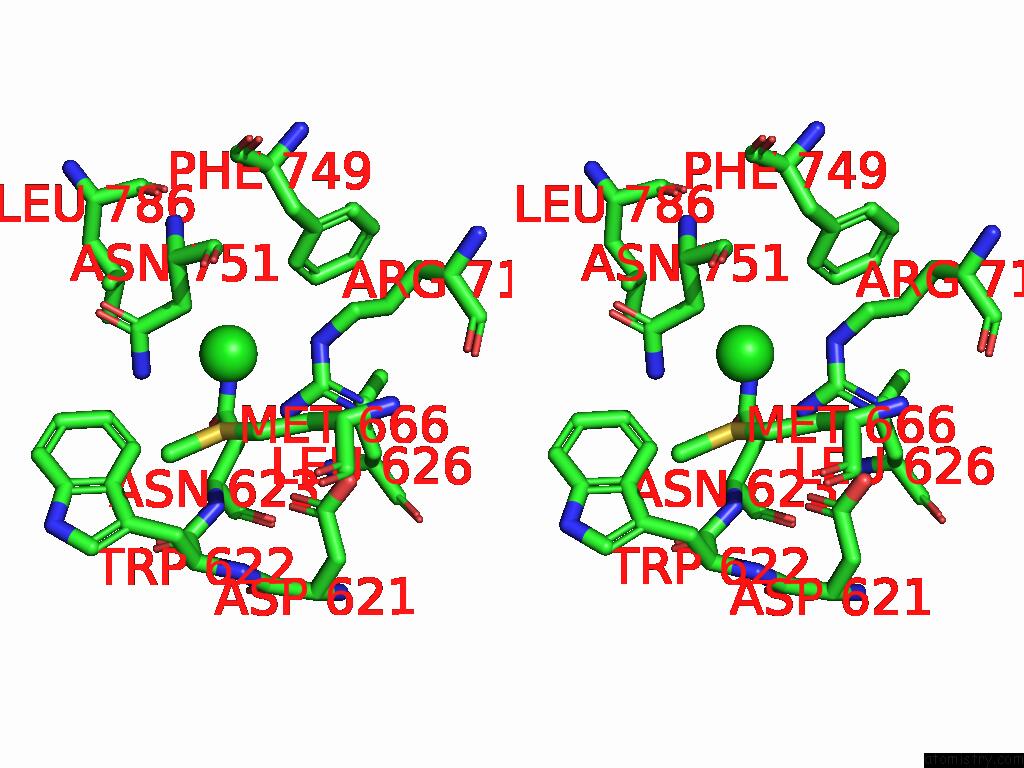
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Cryo-Em Structure of Cellodextrin Phosphorylase From Clostridium Thermocellum with Cellodextrin Ligands within 5.0Å range:
|
Chlorine binding site 2 out of 5 in 8yaq
Go back to
Chlorine binding site 2 out
of 5 in the Cryo-Em Structure of Cellodextrin Phosphorylase From Clostridium Thermocellum with Cellodextrin Ligands
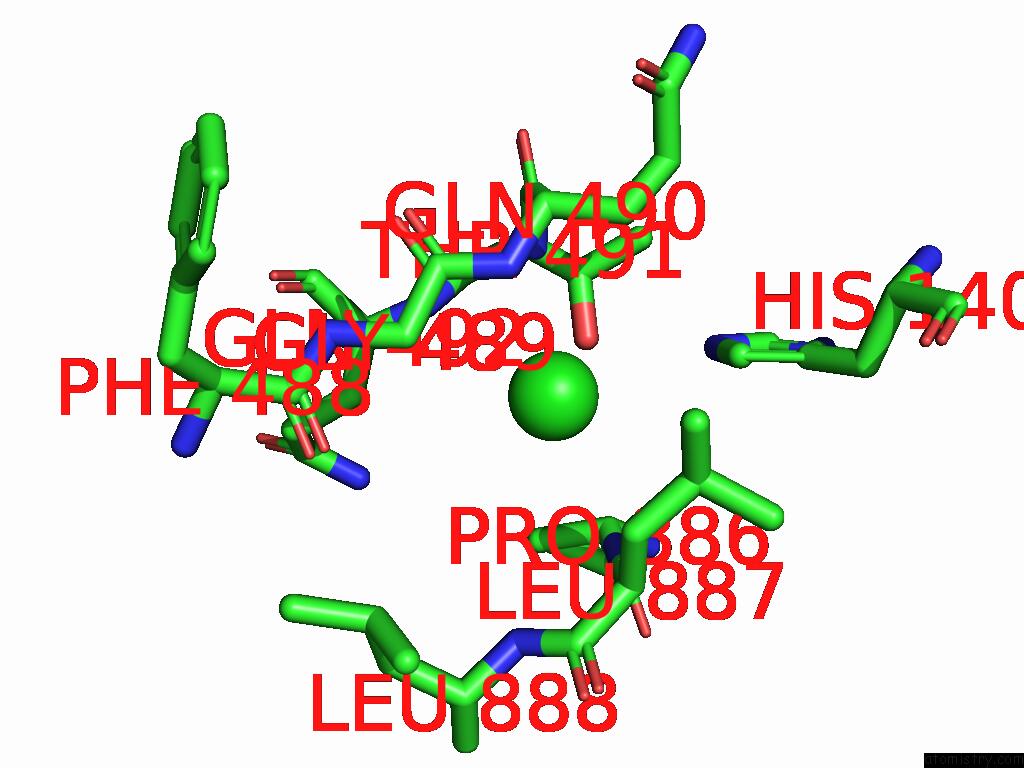
Mono view
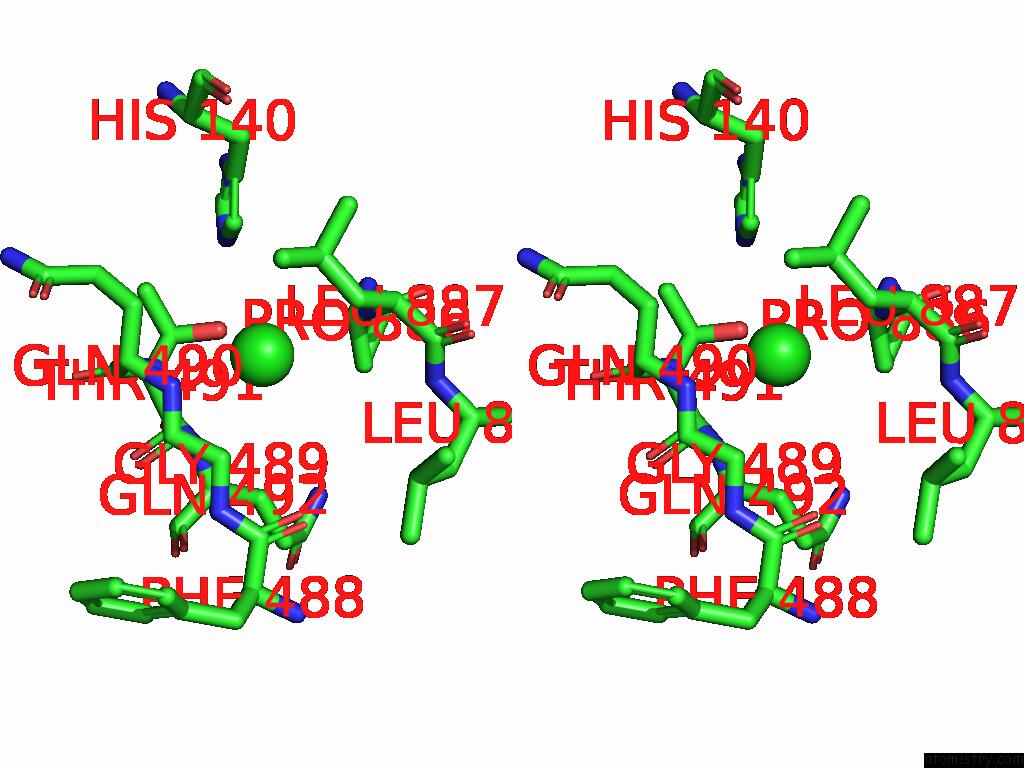
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Cryo-Em Structure of Cellodextrin Phosphorylase From Clostridium Thermocellum with Cellodextrin Ligands within 5.0Å range:
|
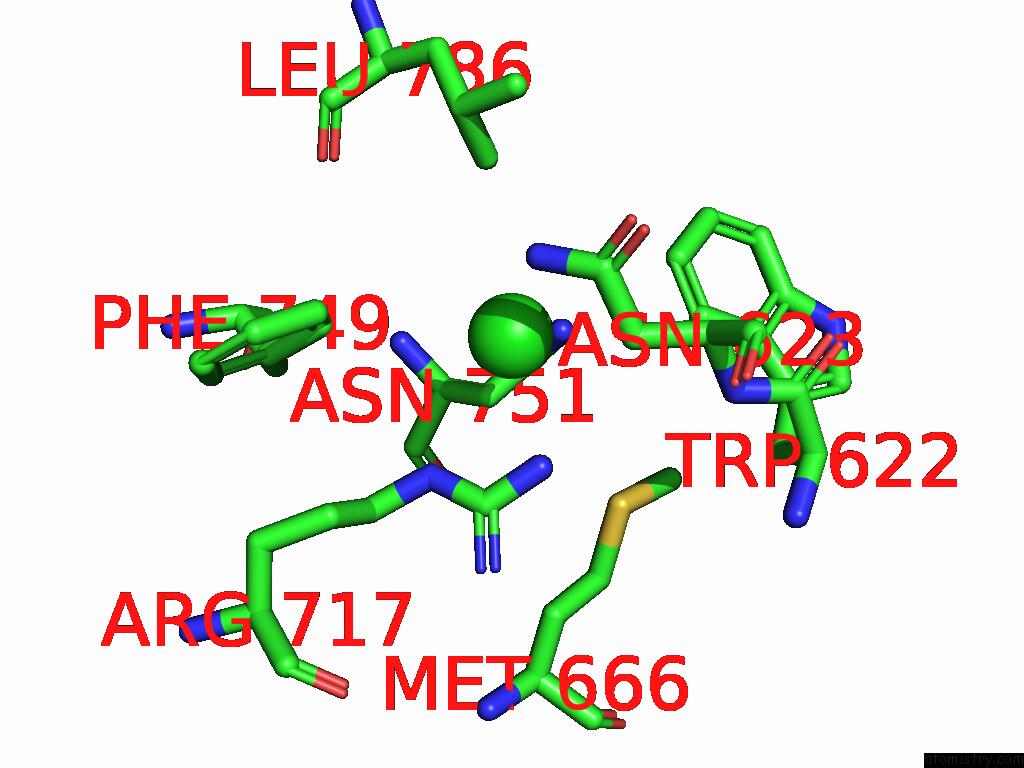
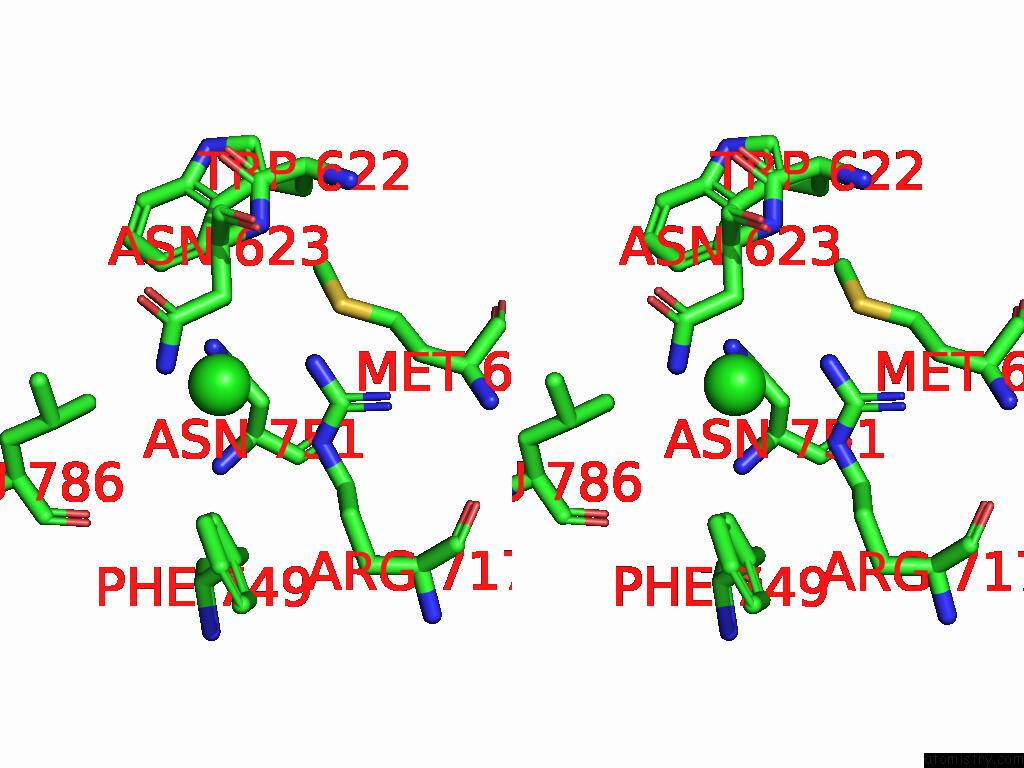
Chlorine binding site 3 out of 5 in 8yaq
Go back to
Chlorine binding site 3 out
of 5 in the Cryo-Em Structure of Cellodextrin Phosphorylase From Clostridium Thermocellum with Cellodextrin Ligands
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Cryo-Em Structure of Cellodextrin Phosphorylase From Clostridium Thermocellum with Cellodextrin Ligands within 5.0Å range:
|
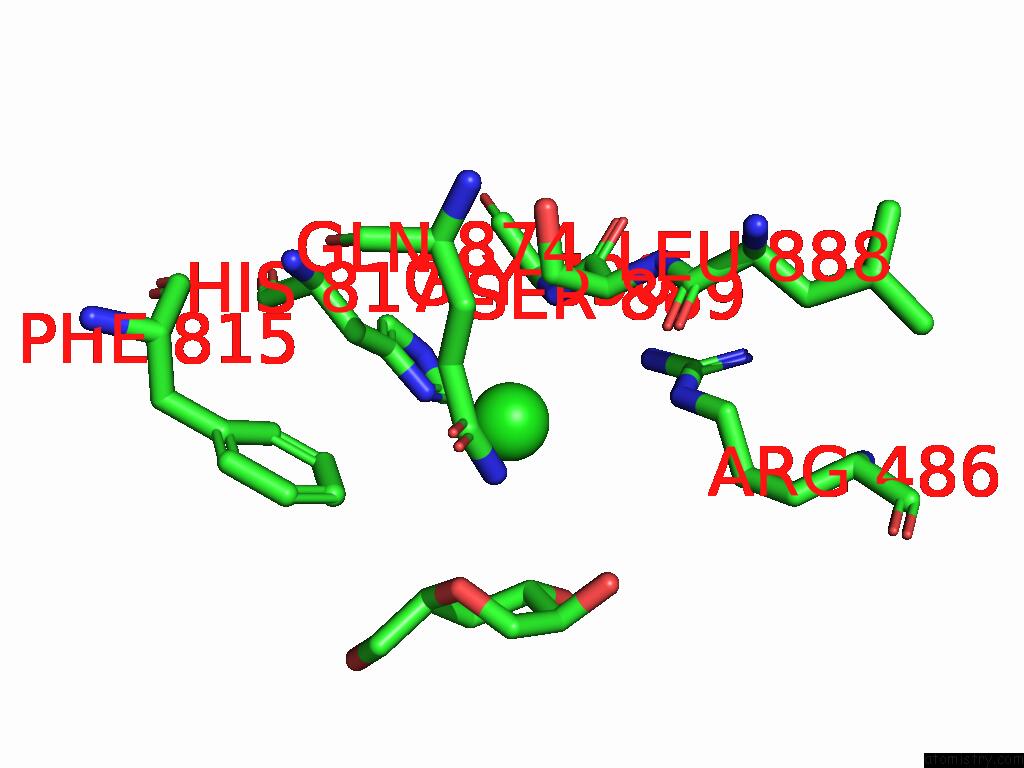
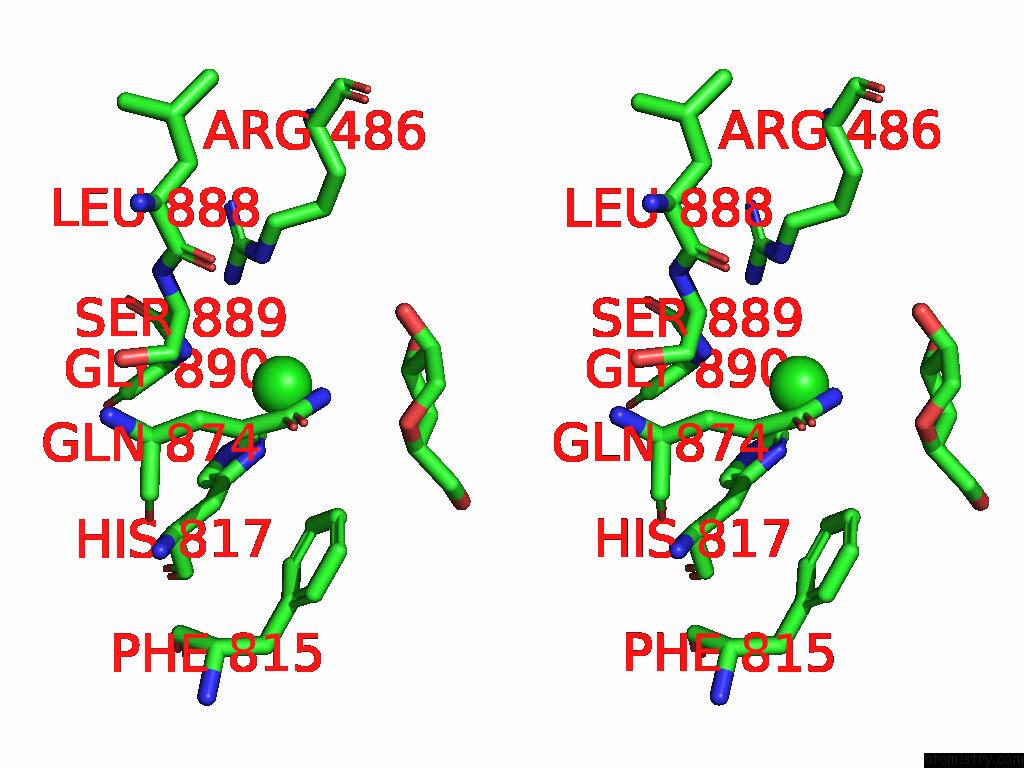
Chlorine binding site 4 out of 5 in 8yaq
Go back to
Chlorine binding site 4 out
of 5 in the Cryo-Em Structure of Cellodextrin Phosphorylase From Clostridium Thermocellum with Cellodextrin Ligands
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Cryo-Em Structure of Cellodextrin Phosphorylase From Clostridium Thermocellum with Cellodextrin Ligands within 5.0Å range:
|
Chlorine binding site 5 out of 5 in 8yaq
Go back to
Chlorine binding site 5 out
of 5 in the Cryo-Em Structure of Cellodextrin Phosphorylase From Clostridium Thermocellum with Cellodextrin Ligands
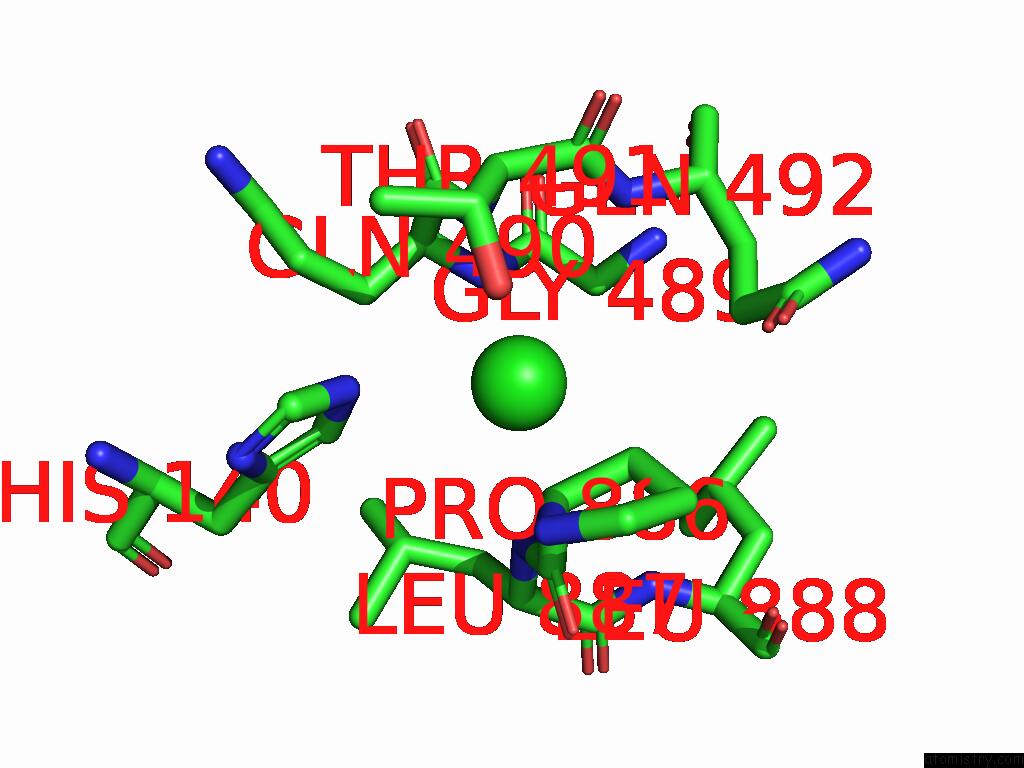
Mono view
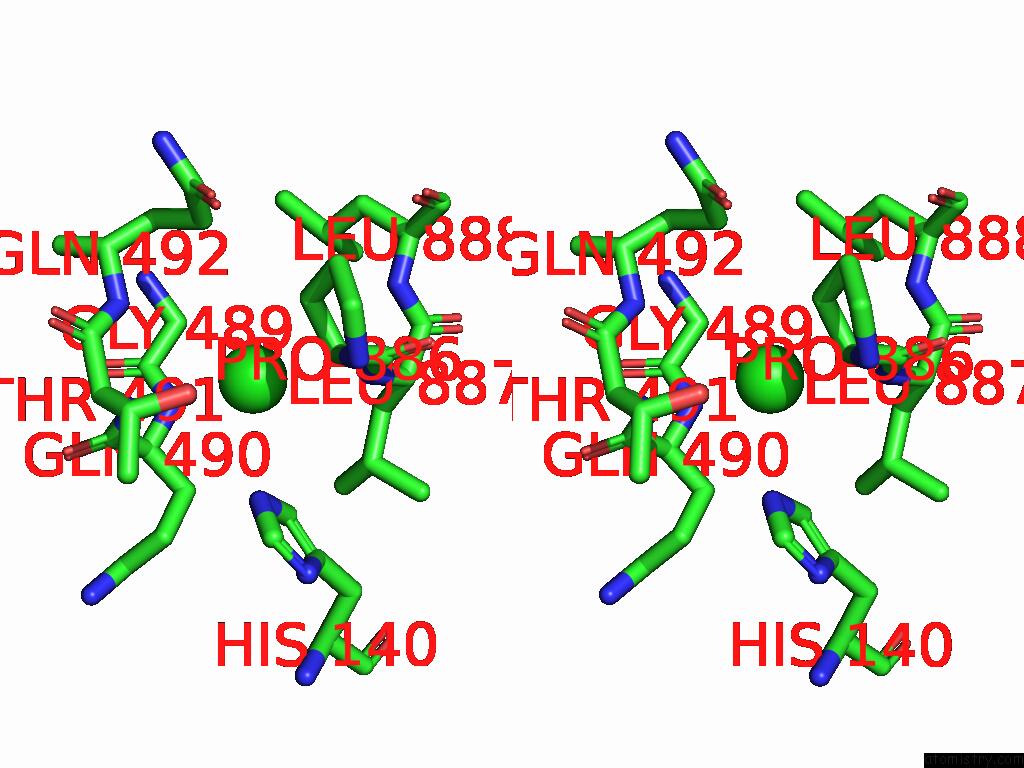
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Cryo-Em Structure of Cellodextrin Phosphorylase From Clostridium Thermocellum with Cellodextrin Ligands within 5.0Å range:
|
Reference:
T.Kuga,
N.Sunagawa,
K.Igarashi.
Structure and Dynamics of Cellodextrin Phosphorylase From Clostridium Thermocellum Determine Chain Length and Crystalline Packing of Highly Ordered Cellulose II Synthesized in Vitro To Be Published.
Page generated: Sun Jul 13 15:45:58 2025
Last articles
I in 9E4XI in 9E50
I in 9DQ1
I in 9DWY
I in 9DAJ
I in 9D9T
I in 9B7C
I in 9AXJ
I in 8Y9X
I in 8ZEV