Chlorine »
PDB 9dpu-9edn »
9dsn »
Chlorine in PDB 9dsn: D306A Mutant of M.Tuberculosis Mend (Sephchc Synthase)
Enzymatic activity of D306A Mutant of M.Tuberculosis Mend (Sephchc Synthase)
All present enzymatic activity of D306A Mutant of M.Tuberculosis Mend (Sephchc Synthase):
2.2.1.9;
2.2.1.9;
Protein crystallography data
The structure of D306A Mutant of M.Tuberculosis Mend (Sephchc Synthase), PDB code: 9dsn
was solved by
J.M.Johnston,
N.A.T.Ho,
F.M.Given,
T.A.Allison,
E.M.M.Bulloch,
W.Jiao,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.75 / 2.30 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 101.217, 139.257, 181.625, 90, 90, 90 |
| R / Rfree (%) | 19.2 / 22.8 |
Other elements in 9dsn:
The structure of D306A Mutant of M.Tuberculosis Mend (Sephchc Synthase) also contains other interesting chemical elements:
| Magnesium | (Mg) | 2 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the D306A Mutant of M.Tuberculosis Mend (Sephchc Synthase)
(pdb code 9dsn). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the D306A Mutant of M.Tuberculosis Mend (Sephchc Synthase), PDB code: 9dsn:
In total only one binding site of Chlorine was determined in the D306A Mutant of M.Tuberculosis Mend (Sephchc Synthase), PDB code: 9dsn:
Chlorine binding site 1 out of 1 in 9dsn
Go back to
Chlorine binding site 1 out
of 1 in the D306A Mutant of M.Tuberculosis Mend (Sephchc Synthase)
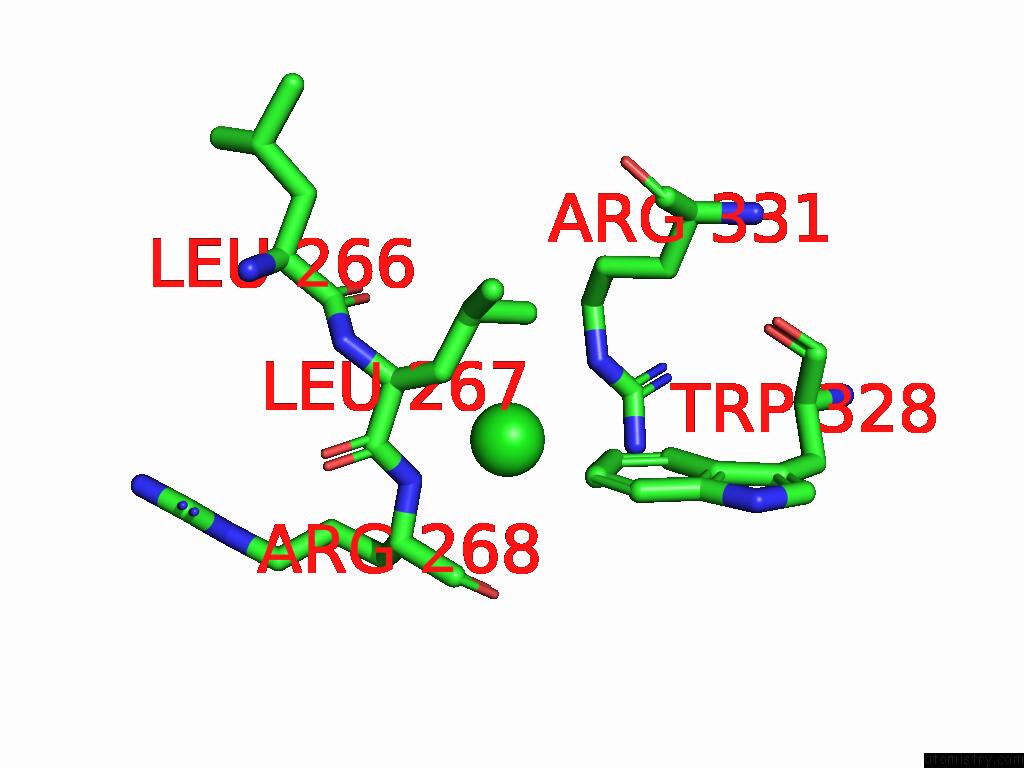
Mono view
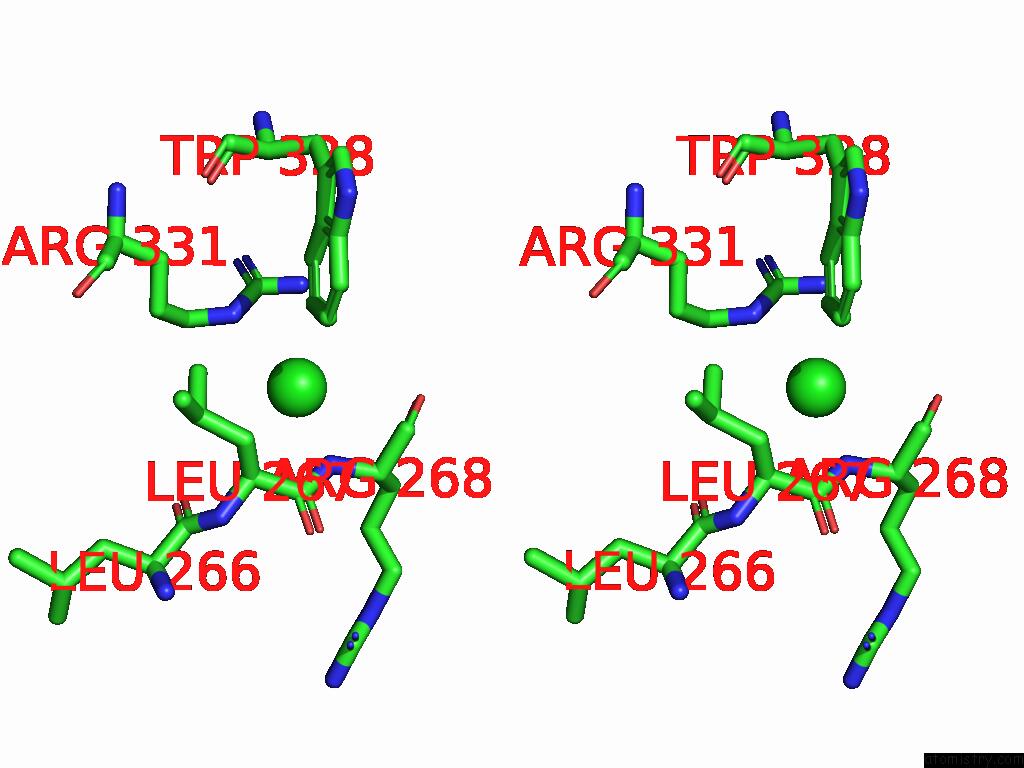
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of D306A Mutant of M.Tuberculosis Mend (Sephchc Synthase) within 5.0Å range:
|
Reference:
N.A.T.Ho,
F.M.Given,
T.Stanborough,
M.Klein,
T.M.Allison,
E.M.M.Bulloch,
W.Jiao,
J.M.Johnston.
Apparent Reversal of Allosteric Response in Mycobacterium Tuberculosis Mend Reveals Links to Half-of-Sites Reactivity. Chembiochem 00943 2025.
ISSN: ESSN 1439-7633
PubMed: 39945237
DOI: 10.1002/CBIC.202400943
Page generated: Sun Jul 13 16:25:29 2025
ISSN: ESSN 1439-7633
PubMed: 39945237
DOI: 10.1002/CBIC.202400943
Last articles
Fe in 4O1ZFe in 4O35
Fe in 4O2G
Fe in 4O1Q
Fe in 4NZI
Fe in 4O1T
Fe in 4NZ2
Fe in 4NY4
Fe in 4NXC
Fe in 4NXA