Chlorine »
PDB 1qhy-1quo »
1quj »
Chlorine in PDB 1quj: Phosphate-Binding Protein Mutant with Asp 137 Replaced By Gly Complex with Chlorine and Phosphate
Protein crystallography data
The structure of Phosphate-Binding Protein Mutant with Asp 137 Replaced By Gly Complex with Chlorine and Phosphate, PDB code: 1quj
was solved by
N.Yao,
A.Choudhary,
P.S.Ledvina,
F.A.Quiocho,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 8.00 / 1.90 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 41.970, 63.980, 123.970, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 19 / 20.9 |
Chlorine Binding Sites:
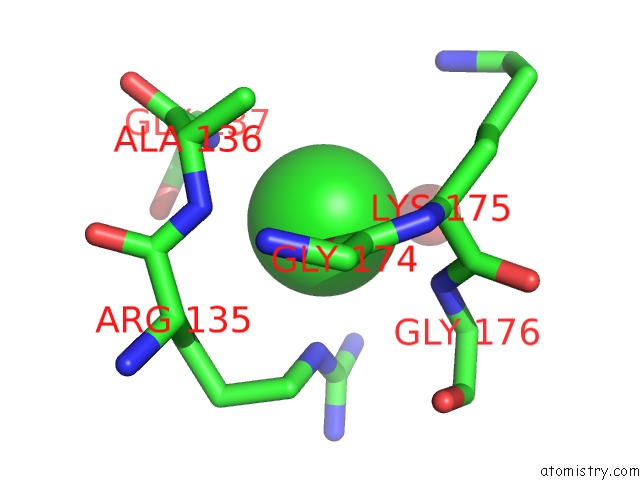
The binding sites of Chlorine atom in the Phosphate-Binding Protein Mutant with Asp 137 Replaced By Gly Complex with Chlorine and Phosphate
(pdb code 1quj). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Phosphate-Binding Protein Mutant with Asp 137 Replaced By Gly Complex with Chlorine and Phosphate, PDB code: 1quj:
In total only one binding site of Chlorine was determined in the Phosphate-Binding Protein Mutant with Asp 137 Replaced By Gly Complex with Chlorine and Phosphate, PDB code: 1quj:
Chlorine binding site 1 out of 1 in 1quj
Go back to
Chlorine binding site 1 out
of 1 in the Phosphate-Binding Protein Mutant with Asp 137 Replaced By Gly Complex with Chlorine and Phosphate
Mono view
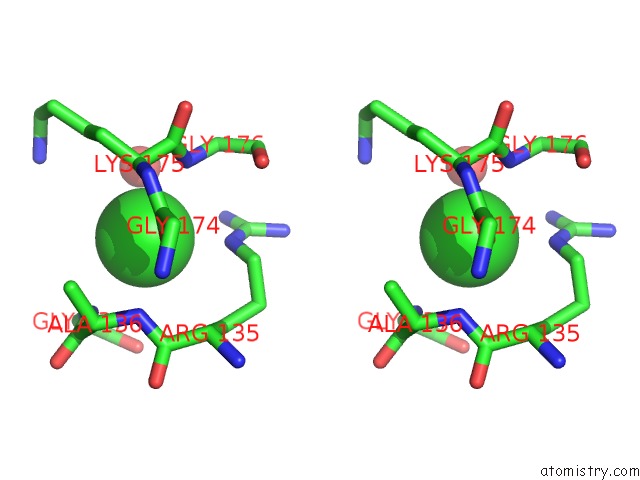
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Phosphate-Binding Protein Mutant with Asp 137 Replaced By Gly Complex with Chlorine and Phosphate within 5.0Å range:
|
Reference:
N.Yao,
P.S.Ledvina,
A.Choudhary,
F.A.Quiocho.
Modulation of A Salt Link Does Not Affect Binding of Phosphate to Its Specific Active Transport Receptor. Biochemistry V. 35 2079 1996.
ISSN: ISSN 0006-2960
PubMed: 8652549
DOI: 10.1021/BI952686R
Page generated: Thu Jul 10 19:08:44 2025
ISSN: ISSN 0006-2960
PubMed: 8652549
DOI: 10.1021/BI952686R
Last articles
Cl in 2VWWCl in 2VWV
Cl in 2VWN
Cl in 2VWU
Cl in 2VWM
Cl in 2VWL
Cl in 2VWD
Cl in 2VVC
Cl in 2VVZ
Cl in 2VVU