Chlorine »
PDB 1wbq-1wvf »
1wut »
Chlorine in PDB 1wut: Acyl Ureas As Human Liver Glycogen Phosphorylase Inhibitors For the Treatment of Type 2 Diabetes
Enzymatic activity of Acyl Ureas As Human Liver Glycogen Phosphorylase Inhibitors For the Treatment of Type 2 Diabetes
All present enzymatic activity of Acyl Ureas As Human Liver Glycogen Phosphorylase Inhibitors For the Treatment of Type 2 Diabetes:
2.4.1.1;
2.4.1.1;
Protein crystallography data
The structure of Acyl Ureas As Human Liver Glycogen Phosphorylase Inhibitors For the Treatment of Type 2 Diabetes, PDB code: 1wut
was solved by
T.Klabunde,
K.U.Wendt,
D.Kadereit,
V.Brachvogel,
H.-J.Burger,
A.W.Herling,
N.G.Oikonomakos,
M.N.Kosmopoulou,
D.Schmoll,
E.Sarubbi,
E.Von Roedern,
K.Schonafinger,
E.Defossa,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.49 / 2.26 |
| Space group | P 43 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 128.679, 128.679, 115.977, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.1 / 23.6 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Acyl Ureas As Human Liver Glycogen Phosphorylase Inhibitors For the Treatment of Type 2 Diabetes
(pdb code 1wut). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 3 binding sites of Chlorine where determined in the Acyl Ureas As Human Liver Glycogen Phosphorylase Inhibitors For the Treatment of Type 2 Diabetes, PDB code: 1wut:
Jump to Chlorine binding site number: 1; 2; 3;
In total 3 binding sites of Chlorine where determined in the Acyl Ureas As Human Liver Glycogen Phosphorylase Inhibitors For the Treatment of Type 2 Diabetes, PDB code: 1wut:
Jump to Chlorine binding site number: 1; 2; 3;
Chlorine binding site 1 out of 3 in 1wut
Go back to
Chlorine binding site 1 out
of 3 in the Acyl Ureas As Human Liver Glycogen Phosphorylase Inhibitors For the Treatment of Type 2 Diabetes
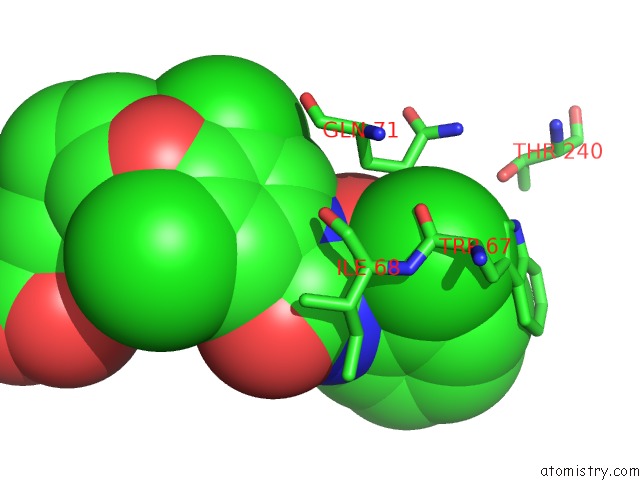
Mono view
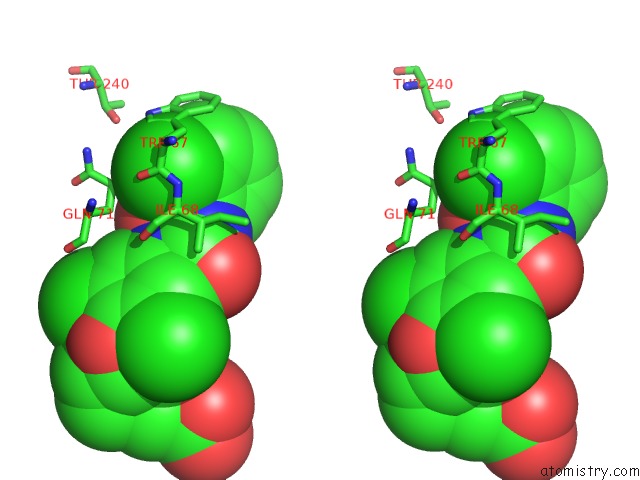
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Acyl Ureas As Human Liver Glycogen Phosphorylase Inhibitors For the Treatment of Type 2 Diabetes within 5.0Å range:
|
Chlorine binding site 2 out of 3 in 1wut
Go back to
Chlorine binding site 2 out
of 3 in the Acyl Ureas As Human Liver Glycogen Phosphorylase Inhibitors For the Treatment of Type 2 Diabetes
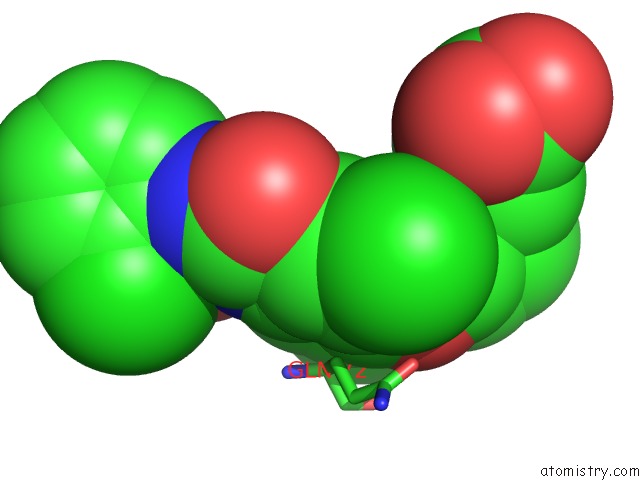
Mono view
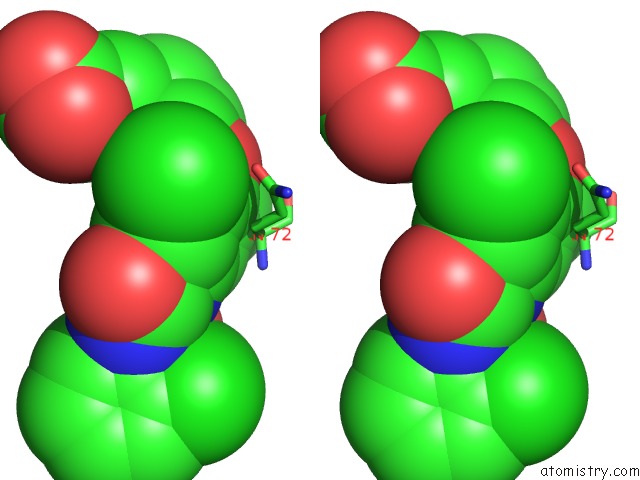
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Acyl Ureas As Human Liver Glycogen Phosphorylase Inhibitors For the Treatment of Type 2 Diabetes within 5.0Å range:
|
Chlorine binding site 3 out of 3 in 1wut
Go back to
Chlorine binding site 3 out
of 3 in the Acyl Ureas As Human Liver Glycogen Phosphorylase Inhibitors For the Treatment of Type 2 Diabetes
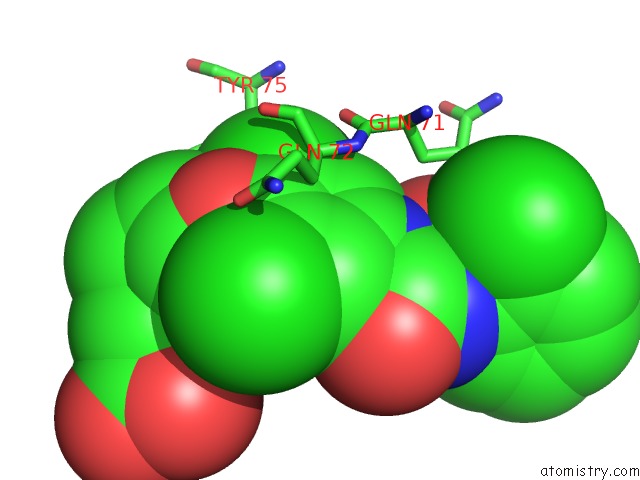
Mono view
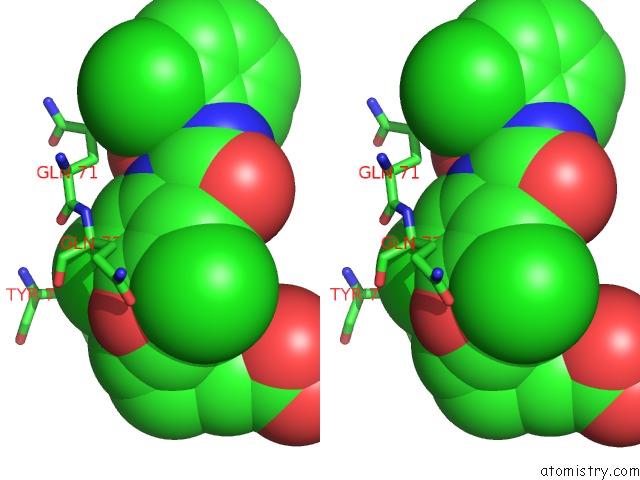
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Acyl Ureas As Human Liver Glycogen Phosphorylase Inhibitors For the Treatment of Type 2 Diabetes within 5.0Å range:
|
Reference:
N.G.Oikonomakos,
M.N.Kosmopoulou,
E.D.Chrysina,
D.D.Leonidas,
I.D.Kostas,
K.U.Wendt,
T.Klabunde,
E.Defossa.
Crystallographic Studies on Acyl Ureas, A New Class of Glycogen Phosphorylase Inhibitors, As Potential Antidiabetic Drugs Protein Sci. V. 14 1760 2005.
ISSN: ISSN 0961-8368
PubMed: 15987904
DOI: 10.1110/PS.051432405
Page generated: Thu Jul 10 20:19:52 2025
ISSN: ISSN 0961-8368
PubMed: 15987904
DOI: 10.1110/PS.051432405
Last articles
Cl in 2WYTCl in 2WZC
Cl in 2WUV
Cl in 2WZB
Cl in 2WYJ
Cl in 2WYG
Cl in 2WVO
Cl in 2WXF
Cl in 2WWP
Cl in 2WW5