Chlorine »
PDB 2kug-2nwh »
2lzg »
Chlorine in PDB 2lzg: uc(Nmr) Structure of MDM2 (6-125) with Pip-1
Chlorine Binding Sites:
The binding sites of Chlorine atom in the uc(Nmr) Structure of MDM2 (6-125) with Pip-1
(pdb code 2lzg). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the uc(Nmr) Structure of MDM2 (6-125) with Pip-1, PDB code: 2lzg:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the uc(Nmr) Structure of MDM2 (6-125) with Pip-1, PDB code: 2lzg:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 2lzg
Go back to
Chlorine binding site 1 out
of 2 in the uc(Nmr) Structure of MDM2 (6-125) with Pip-1
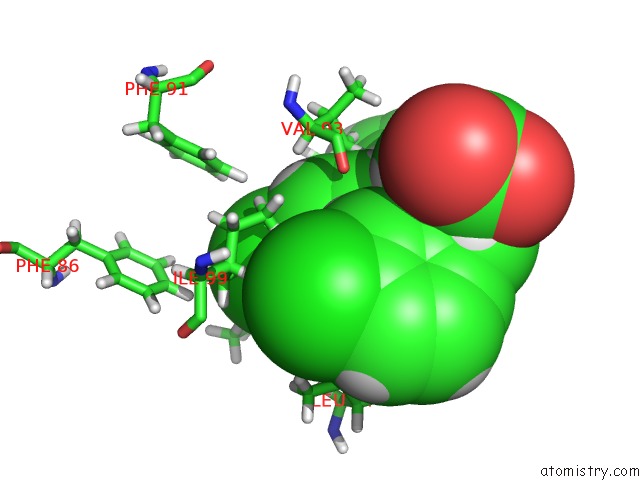
Mono view
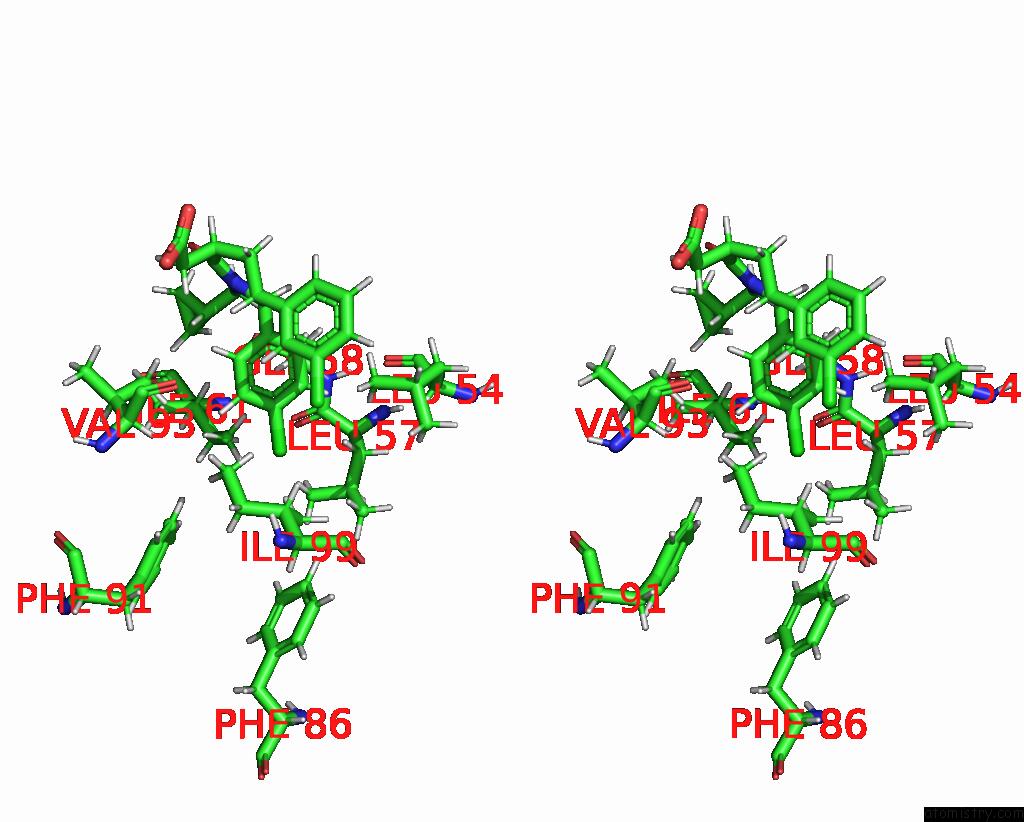
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of uc(Nmr) Structure of MDM2 (6-125) with Pip-1 within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 2lzg
Go back to
Chlorine binding site 2 out
of 2 in the uc(Nmr) Structure of MDM2 (6-125) with Pip-1
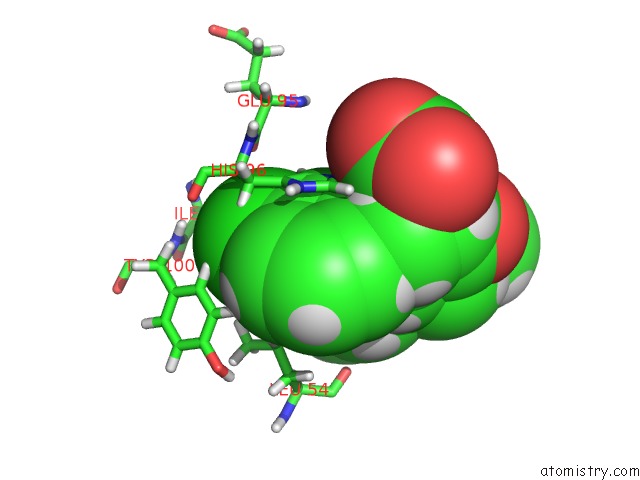
Mono view
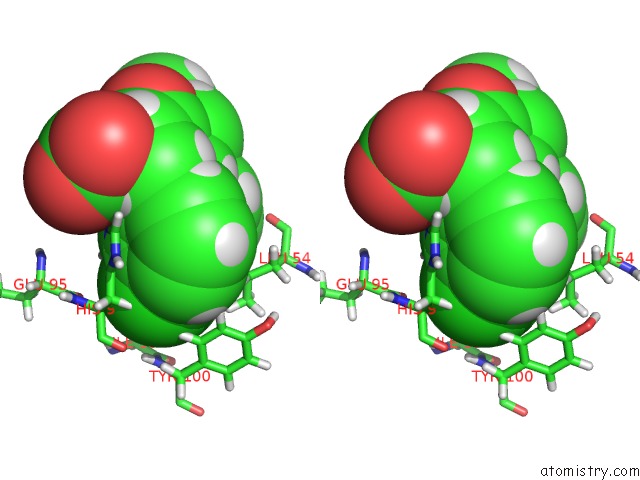
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of uc(Nmr) Structure of MDM2 (6-125) with Pip-1 within 5.0Å range:
|
Reference:
K.Michelsen,
J.B.Jordan,
J.Lewis,
A.M.Long,
E.Yang,
Y.Rew,
J.Zhou,
P.Yakowec,
P.D.Schnier,
X.Huang,
L.Poppe.
Ordering of the N-Terminus of Human MDM2 By Small Molecule Inhibitors. J.Am.Chem.Soc. V. 134 17059 2012.
ISSN: ISSN 0002-7863
PubMed: 22991965
DOI: 10.1021/JA305839B
Page generated: Thu Jul 10 23:17:19 2025
ISSN: ISSN 0002-7863
PubMed: 22991965
DOI: 10.1021/JA305839B
Last articles
Cl in 3E5UCl in 3E05
Cl in 3E3K
Cl in 3E4B
Cl in 3E15
Cl in 3E1T
Cl in 3E18
Cl in 3E0R
Cl in 3DZU
Cl in 3DZZ