Chlorine »
PDB 2xmd-2xs6 »
2xs6 »
Chlorine in PDB 2xs6: Crystal Structure of the Rhogap Domain of Human PIK3R2
Protein crystallography data
The structure of Crystal Structure of the Rhogap Domain of Human PIK3R2, PDB code: 2xs6
was solved by
L.Tresaugues,
M.Welin,
C.H.Arrowsmith,
H.Berglund,
C.Bountra,
R.Collins,
A.M.Edwards,
S.Flodin,
A.Flores,
S.Graslund,
M.Hammarstrom,
I.Johansson,
T.Karlberg,
S.Kol,
T.Kotenyova,
E.Kouznetsova,
M.Moche,
T.Nyman,
C.Persson,
H.Schuler,
P.Schutz,
M.I.Siponen,
A.G.Thorsell,
S.Van Der Berg,
E.Wahlberg,
J.Weigelt,
P.Nordlund,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 53.92 / 2.09 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 94.661, 34.354, 71.317, 90.00, 130.88, 90.00 |
| R / Rfree (%) | 19.149 / 23.98 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of the Rhogap Domain of Human PIK3R2
(pdb code 2xs6). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Crystal Structure of the Rhogap Domain of Human PIK3R2, PDB code: 2xs6:
In total only one binding site of Chlorine was determined in the Crystal Structure of the Rhogap Domain of Human PIK3R2, PDB code: 2xs6:
Chlorine binding site 1 out of 1 in 2xs6
Go back to
Chlorine binding site 1 out
of 1 in the Crystal Structure of the Rhogap Domain of Human PIK3R2
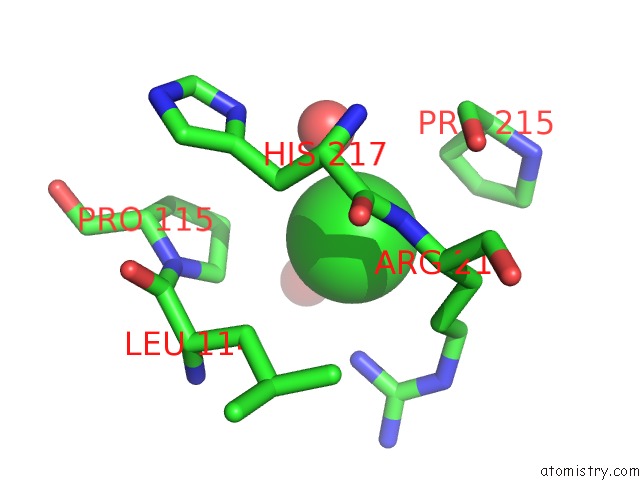
Mono view
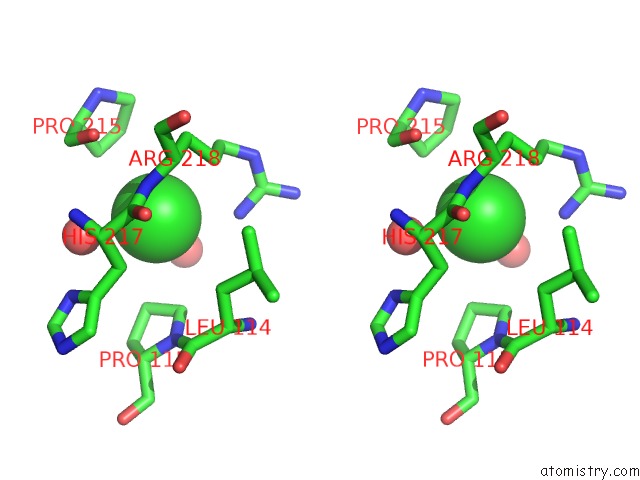
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of the Rhogap Domain of Human PIK3R2 within 5.0Å range:
|
Reference:
S.M.Lim,
K.Yeung,
L.Tresaugues,
T.H.Ling,
P.Nordlund.
The Structure and Catalytic Mechanism of Human Sphingomyelin Phosphodiesterase Like 3A - An Acid Sphingomyelinase Homolog with A Novel Nucleotide Hydrolase Activity. Febs J. V. 283 1107 2016.
ISSN: ISSN 1742-464X
PubMed: 26783088
DOI: 10.1111/FEBS.13655
Page generated: Fri Jul 11 02:07:37 2025
ISSN: ISSN 1742-464X
PubMed: 26783088
DOI: 10.1111/FEBS.13655
Last articles
Cl in 3OZDCl in 3OZR
Cl in 3OYZ
Cl in 3OYX
Cl in 3OZ0
Cl in 3OZC
Cl in 3OYO
Cl in 3OYV
Cl in 3OXO
Cl in 3OYL