Chlorine »
PDB 3b0x-3bft »
3bd1 »
Chlorine in PDB 3bd1: Structure of the Cro Protein From Putative Prophage Element Xfaso 1 in Xylella Fastidiosa Strain Ann-1
Protein crystallography data
The structure of Structure of the Cro Protein From Putative Prophage Element Xfaso 1 in Xylella Fastidiosa Strain Ann-1, PDB code: 3bd1
was solved by
B.M.Hall,
S.A.Roberts,
W.R.Montfort,
M.H.Cordes,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 17.06 / 1.40 |
| Space group | P 32 |
| Cell size a, b, c (Å), α, β, γ (°) | 66.970, 66.970, 54.360, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 14.7 / 18.2 |
Chlorine Binding Sites:
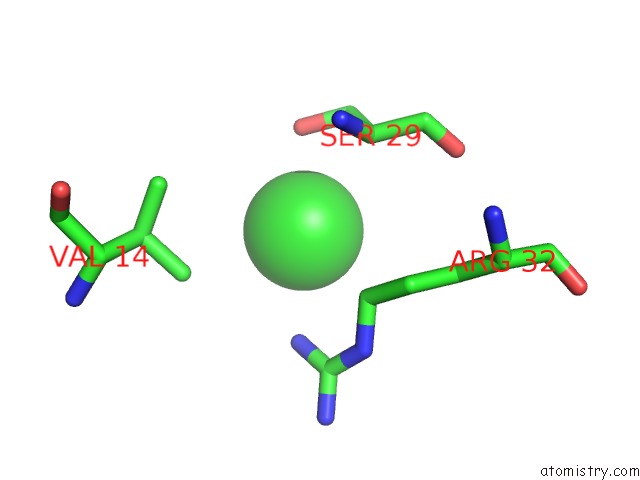
The binding sites of Chlorine atom in the Structure of the Cro Protein From Putative Prophage Element Xfaso 1 in Xylella Fastidiosa Strain Ann-1
(pdb code 3bd1). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Structure of the Cro Protein From Putative Prophage Element Xfaso 1 in Xylella Fastidiosa Strain Ann-1, PDB code: 3bd1:
In total only one binding site of Chlorine was determined in the Structure of the Cro Protein From Putative Prophage Element Xfaso 1 in Xylella Fastidiosa Strain Ann-1, PDB code: 3bd1:
Chlorine binding site 1 out of 1 in 3bd1
Go back to
Chlorine binding site 1 out
of 1 in the Structure of the Cro Protein From Putative Prophage Element Xfaso 1 in Xylella Fastidiosa Strain Ann-1
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Structure of the Cro Protein From Putative Prophage Element Xfaso 1 in Xylella Fastidiosa Strain Ann-1 within 5.0Å range:
|
Reference:
C.G.Roessler,
B.M.Hall,
W.J.Anderson,
W.M.Ingram,
S.A.Roberts,
W.R.Montfort,
M.H.Cordes.
Transitive Homology-Guided Structural Studies Lead to Discovery of Cro Proteins with 40% Sequence Identity But Different Folds Proc.Natl.Acad.Sci.Usa V. 105 2343 2008.
ISSN: ISSN 0027-8424
PubMed: 18227506
DOI: 10.1073/PNAS.0711589105
Page generated: Fri Jul 11 03:25:23 2025
ISSN: ISSN 0027-8424
PubMed: 18227506
DOI: 10.1073/PNAS.0711589105
Last articles
Cl in 3ON5Cl in 3OMN
Cl in 3OMM
Cl in 3OMK
Cl in 3OMI
Cl in 3OLZ
Cl in 3OLW
Cl in 3OMC
Cl in 3OLF
Cl in 3OLI