Chlorine »
PDB 3d4i-3dgo »
3dee »
Chlorine in PDB 3dee: Crystal Structure of A Putative Regulatory Protein Involved in Transcription (NGO1945) From Neisseria Gonorrhoeae Fa 1090 at 2.25 A Resolution
Protein crystallography data
The structure of Crystal Structure of A Putative Regulatory Protein Involved in Transcription (NGO1945) From Neisseria Gonorrhoeae Fa 1090 at 2.25 A Resolution, PDB code: 3dee
was solved by
Joint Center For Structural Genomics (Jcsg),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 77.85 / 2.10 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 106.522, 31.879, 86.369, 90.00, 115.80, 90.00 |
| R / Rfree (%) | 22.3 / 26.7 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of A Putative Regulatory Protein Involved in Transcription (NGO1945) From Neisseria Gonorrhoeae Fa 1090 at 2.25 A Resolution
(pdb code 3dee). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Crystal Structure of A Putative Regulatory Protein Involved in Transcription (NGO1945) From Neisseria Gonorrhoeae Fa 1090 at 2.25 A Resolution, PDB code: 3dee:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Crystal Structure of A Putative Regulatory Protein Involved in Transcription (NGO1945) From Neisseria Gonorrhoeae Fa 1090 at 2.25 A Resolution, PDB code: 3dee:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 3dee
Go back to
Chlorine binding site 1 out
of 2 in the Crystal Structure of A Putative Regulatory Protein Involved in Transcription (NGO1945) From Neisseria Gonorrhoeae Fa 1090 at 2.25 A Resolution
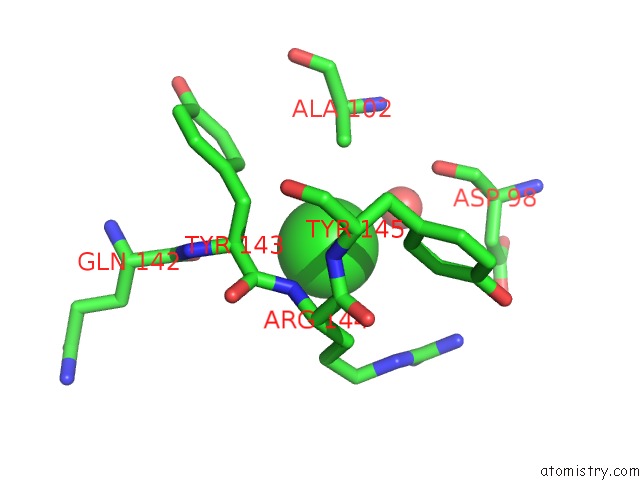
Mono view
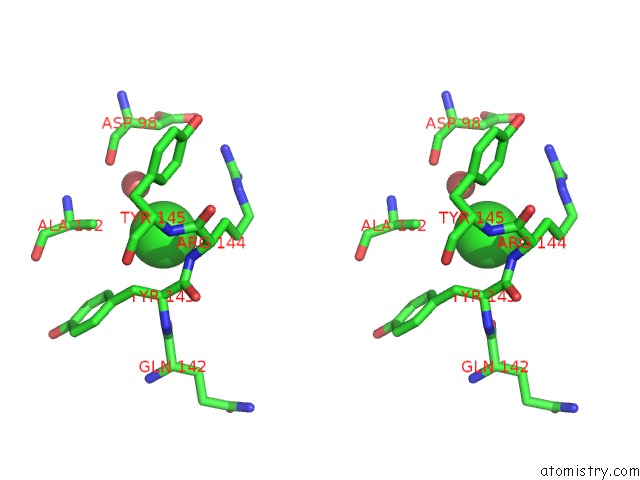
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of A Putative Regulatory Protein Involved in Transcription (NGO1945) From Neisseria Gonorrhoeae Fa 1090 at 2.25 A Resolution within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 3dee
Go back to
Chlorine binding site 2 out
of 2 in the Crystal Structure of A Putative Regulatory Protein Involved in Transcription (NGO1945) From Neisseria Gonorrhoeae Fa 1090 at 2.25 A Resolution
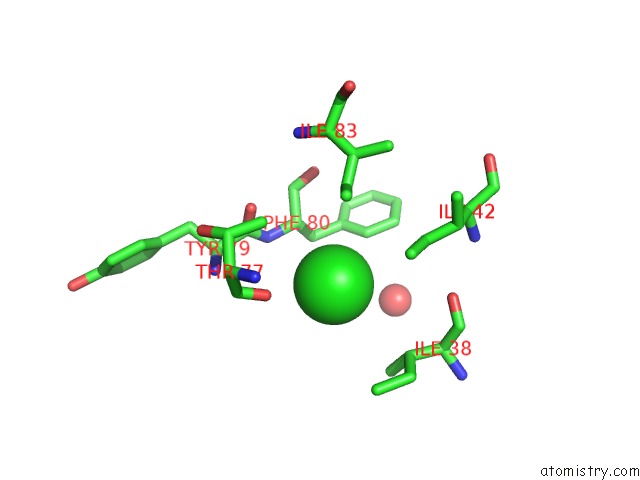
Mono view
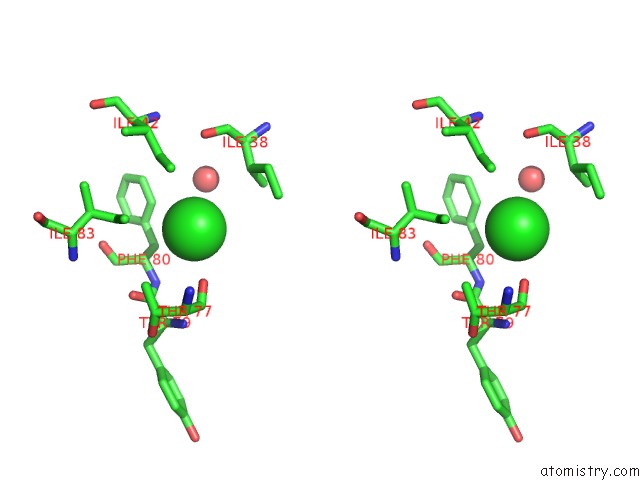
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of A Putative Regulatory Protein Involved in Transcription (NGO1945) From Neisseria Gonorrhoeae Fa 1090 at 2.25 A Resolution within 5.0Å range:
|
Reference:
D.Das,
N.V.Grishin,
A.Kumar,
D.Carlton,
C.Bakolitsa,
M.D.Miller,
P.Abdubek,
T.Astakhova,
H.L.Axelrod,
P.Burra,
C.Chen,
H.J.Chiu,
M.Chiu,
T.Clayton,
M.C.Deller,
L.Duan,
K.Ellrott,
D.Ernst,
C.L.Farr,
J.Feuerhelm,
A.Grzechnik,
S.K.Grzechnik,
J.C.Grant,
G.W.Han,
L.Jaroszewski,
K.K.Jin,
H.A.Johnson,
H.E.Klock,
M.W.Knuth,
P.Kozbial,
S.S.Krishna,
D.Marciano,
D.Mcmullan,
A.T.Morse,
E.Nigoghossian,
A.Nopakun,
L.Okach,
S.Oommachen,
J.Paulsen,
C.Puckett,
R.Reyes,
C.L.Rife,
N.Sefcovic,
H.J.Tien,
C.B.Trame,
H.Van Den Bedem,
D.Weekes,
T.Wooten,
Q.Xu,
K.O.Hodgson,
J.Wooley,
M.A.Elsliger,
A.M.Deacon,
A.Godzik,
S.A.Lesley,
I.A.Wilson.
The Structure of the First Representative of Pfam Family PF09836 Reveals A Two-Domain Organization and Suggests Involvement in Transcriptional Regulation. Acta Crystallogr.,Sect.F V. 66 1174 2010.
ISSN: ESSN 1744-3091
PubMed: 20944208
DOI: 10.1107/S1744309109022672
Page generated: Fri Jul 11 04:26:58 2025
ISSN: ESSN 1744-3091
PubMed: 20944208
DOI: 10.1107/S1744309109022672
Last articles
Cl in 3VBFCl in 3VAQ
Cl in 3VAS
Cl in 3VAV
Cl in 3VA9
Cl in 3V91
Cl in 3V90
Cl in 3V7J
Cl in 3V8Z
Cl in 3V7L