Chlorine »
PDB 3itd-3jpy »
3ite »
Chlorine in PDB 3ite: The Third Adenylation Domain of the Fungal Sidn Non-Ribosomal Peptide Synthetase
Protein crystallography data
The structure of The Third Adenylation Domain of the Fungal Sidn Non-Ribosomal Peptide Synthetase, PDB code: 3ite
was solved by
T.V.Lee,
J.S.Lott,
R.D.Johnson,
L.J.Johnson,
V.L.Arcus,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 42.62 / 2.00 |
| Space group | P 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 50.598, 75.280, 84.137, 114.85, 94.78, 90.18 |
| R / Rfree (%) | 19.4 / 22.8 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the The Third Adenylation Domain of the Fungal Sidn Non-Ribosomal Peptide Synthetase
(pdb code 3ite). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the The Third Adenylation Domain of the Fungal Sidn Non-Ribosomal Peptide Synthetase, PDB code: 3ite:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the The Third Adenylation Domain of the Fungal Sidn Non-Ribosomal Peptide Synthetase, PDB code: 3ite:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 3ite
Go back to
Chlorine binding site 1 out
of 2 in the The Third Adenylation Domain of the Fungal Sidn Non-Ribosomal Peptide Synthetase
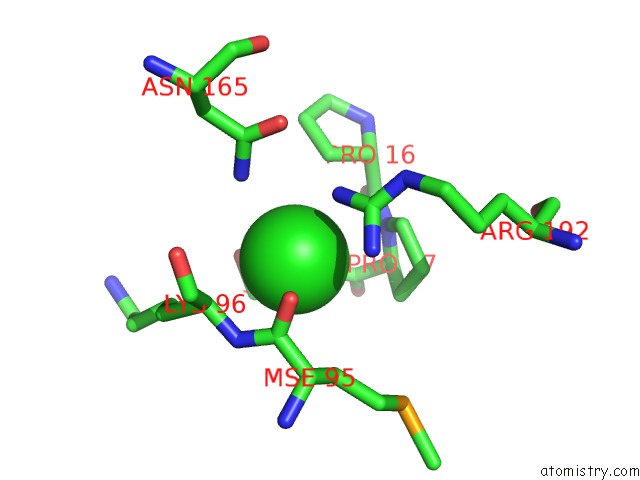
Mono view
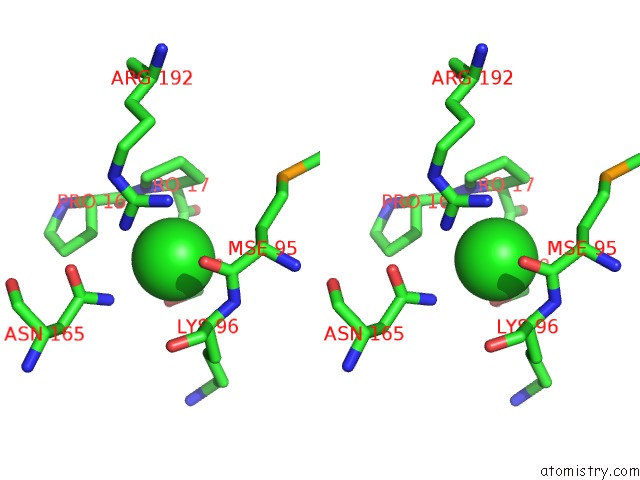
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of The Third Adenylation Domain of the Fungal Sidn Non-Ribosomal Peptide Synthetase within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 3ite
Go back to
Chlorine binding site 2 out
of 2 in the The Third Adenylation Domain of the Fungal Sidn Non-Ribosomal Peptide Synthetase
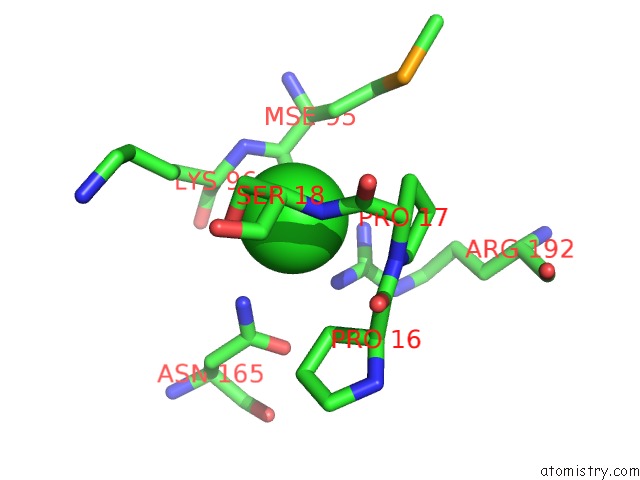
Mono view
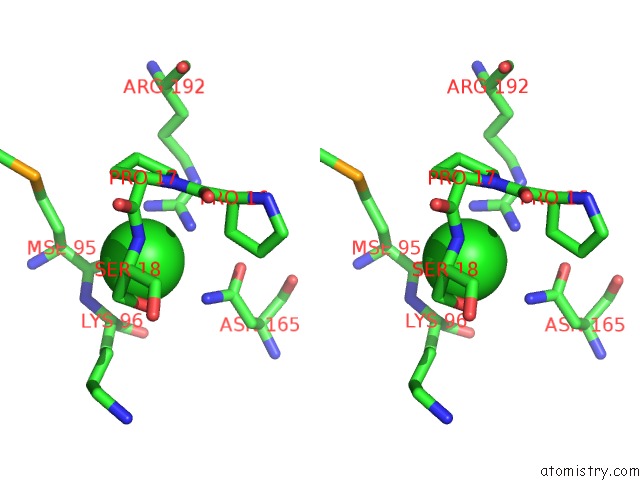
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of The Third Adenylation Domain of the Fungal Sidn Non-Ribosomal Peptide Synthetase within 5.0Å range:
|
Reference:
T.V.Lee,
L.J.Johnson,
R.D.Johnson,
A.Koulman,
G.A.Lane,
J.S.Lott,
V.L.Arcus.
Structure of A Eukaryotic Nonribosomal Peptide Synthetase Adenylation Domain That Activates A Large Hydroxamate Amino Acid in Siderophore Biosynthesis J.Biol.Chem. V. 285 2415 2010.
ISSN: ISSN 0021-9258
PubMed: 19923209
DOI: 10.1074/JBC.M109.071324
Page generated: Fri Jul 11 06:35:43 2025
ISSN: ISSN 0021-9258
PubMed: 19923209
DOI: 10.1074/JBC.M109.071324
Last articles
Cl in 5T67Cl in 5T5X
Cl in 5T5P
Cl in 5T4S
Cl in 5T55
Cl in 5T5L
Cl in 5T4N
Cl in 5T45
Cl in 5T3N
Cl in 5T4M