Chlorine »
PDB 3k9u-3khi »
3kbk »
Chlorine in PDB 3kbk: Epi-Isozizaene Synthase Complexed with Hg
Enzymatic activity of Epi-Isozizaene Synthase Complexed with Hg
All present enzymatic activity of Epi-Isozizaene Synthase Complexed with Hg:
4.2.3.37;
4.2.3.37;
Protein crystallography data
The structure of Epi-Isozizaene Synthase Complexed with Hg, PDB code: 3kbk
was solved by
J.A.Aaron,
X.Lin,
D.E.Cane,
D.W.Christianson,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 37.74 / 1.90 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 51.693, 46.549, 75.551, 90.00, 92.34, 90.00 |
| R / Rfree (%) | 16.9 / 20.2 |
Other elements in 3kbk:
The structure of Epi-Isozizaene Synthase Complexed with Hg also contains other interesting chemical elements:
| Mercury | (Hg) | 6 atoms |
| Sodium | (Na) | 1 atom |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Epi-Isozizaene Synthase Complexed with Hg
(pdb code 3kbk). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 3 binding sites of Chlorine where determined in the Epi-Isozizaene Synthase Complexed with Hg, PDB code: 3kbk:
Jump to Chlorine binding site number: 1; 2; 3;
In total 3 binding sites of Chlorine where determined in the Epi-Isozizaene Synthase Complexed with Hg, PDB code: 3kbk:
Jump to Chlorine binding site number: 1; 2; 3;
Chlorine binding site 1 out of 3 in 3kbk
Go back to
Chlorine binding site 1 out
of 3 in the Epi-Isozizaene Synthase Complexed with Hg

Mono view
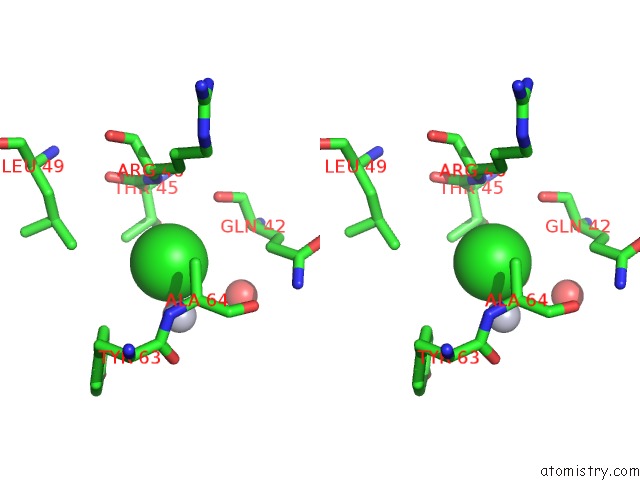
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Epi-Isozizaene Synthase Complexed with Hg within 5.0Å range:
|
Chlorine binding site 2 out of 3 in 3kbk
Go back to
Chlorine binding site 2 out
of 3 in the Epi-Isozizaene Synthase Complexed with Hg
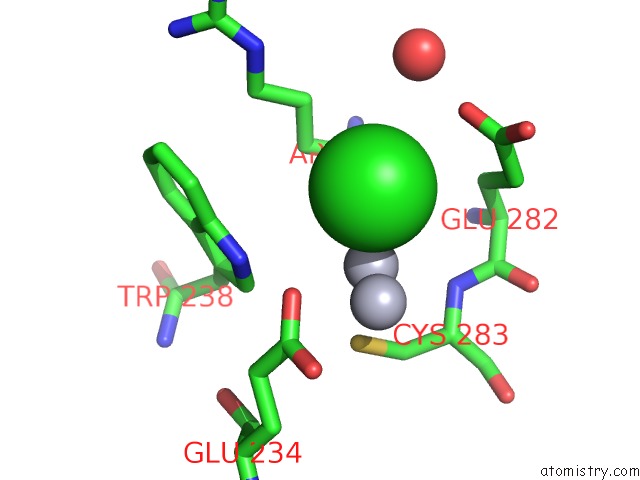
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Epi-Isozizaene Synthase Complexed with Hg within 5.0Å range:
|
Chlorine binding site 3 out of 3 in 3kbk
Go back to
Chlorine binding site 3 out
of 3 in the Epi-Isozizaene Synthase Complexed with Hg

Mono view
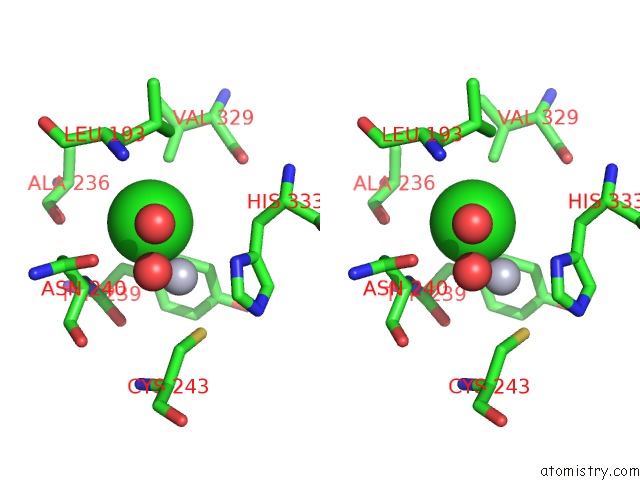
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Epi-Isozizaene Synthase Complexed with Hg within 5.0Å range:
|
Reference:
J.A.Aaron,
X.Lin,
D.E.Cane,
D.W.Christianson.
Structure of Epi-Isozizaene Synthase From Streptomyces Coelicolor A3(2), A Platform For New Terpenoid Cyclization Templates Biochemistry V. 49 1787 2010.
ISSN: ISSN 0006-2960
PubMed: 20131801
DOI: 10.1021/BI902088Z
Page generated: Fri Jul 11 06:56:13 2025
ISSN: ISSN 0006-2960
PubMed: 20131801
DOI: 10.1021/BI902088Z
Last articles
Cl in 5URQCl in 5UTN
Cl in 5USS
Cl in 5URU
Cl in 5USQ
Cl in 5UQH
Cl in 5UQG
Cl in 5UR1
Cl in 5UQS
Cl in 5UQP