Chlorine »
PDB 3rv4-3s8n »
3s1u »
Chlorine in PDB 3s1u: Transaldolase From Thermoplasma Acidophilum in Complex with D- Erythrose 4-Phosphate
Enzymatic activity of Transaldolase From Thermoplasma Acidophilum in Complex with D- Erythrose 4-Phosphate
All present enzymatic activity of Transaldolase From Thermoplasma Acidophilum in Complex with D- Erythrose 4-Phosphate:
2.2.1.2;
2.2.1.2;
Protein crystallography data
The structure of Transaldolase From Thermoplasma Acidophilum in Complex with D- Erythrose 4-Phosphate, PDB code: 3s1u
was solved by
A.Lehwess-Litzmann,
P.Neumann,
C.Parthier,
K.Tittmann,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 19.85 / 1.90 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 148.200, 171.400, 99.100, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.5 / 19.9 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Transaldolase From Thermoplasma Acidophilum in Complex with D- Erythrose 4-Phosphate
(pdb code 3s1u). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Transaldolase From Thermoplasma Acidophilum in Complex with D- Erythrose 4-Phosphate, PDB code: 3s1u:
In total only one binding site of Chlorine was determined in the Transaldolase From Thermoplasma Acidophilum in Complex with D- Erythrose 4-Phosphate, PDB code: 3s1u:
Chlorine binding site 1 out of 1 in 3s1u
Go back to
Chlorine binding site 1 out
of 1 in the Transaldolase From Thermoplasma Acidophilum in Complex with D- Erythrose 4-Phosphate
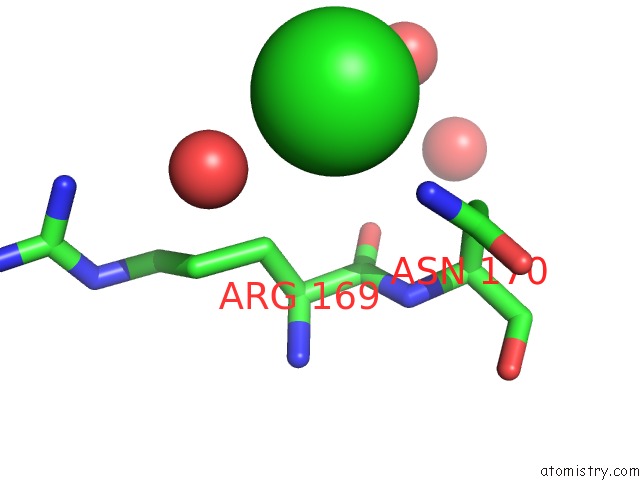
Mono view
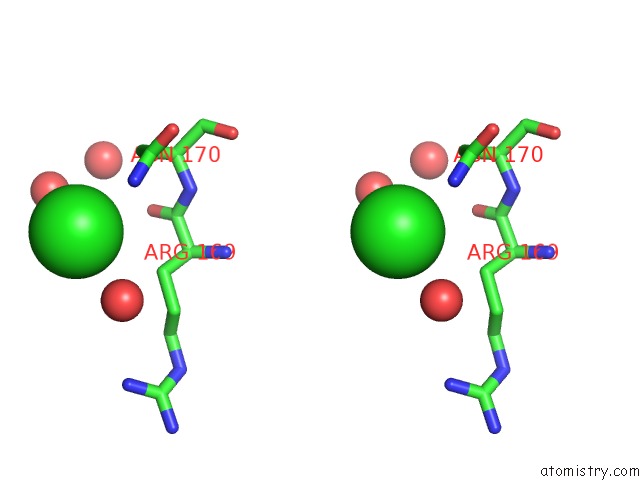
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Transaldolase From Thermoplasma Acidophilum in Complex with D- Erythrose 4-Phosphate within 5.0Å range:
|
Reference:
A.Lehwess-Litzmann,
P.Neumann,
C.Parthier,
S.Ludtke,
R.Golbik,
R.Ficner,
K.Tittmann.
Twisted Schiff Base Intermediates and Substrate Locale Revise Transaldolase Mechanism. Nat.Chem.Biol. V. 7 678 2011.
ISSN: ISSN 1552-4450
PubMed: 21857661
DOI: 10.1038/NCHEMBIO.633
Page generated: Fri Jul 11 10:04:45 2025
ISSN: ISSN 1552-4450
PubMed: 21857661
DOI: 10.1038/NCHEMBIO.633
Last articles
Cl in 4E4JCl in 4E8Y
Cl in 4E7V
Cl in 4E8S
Cl in 4E86
Cl in 4E84
Cl in 4E7R
Cl in 4E83
Cl in 4E69
Cl in 4E6X