Chlorine »
PDB 3zx3-4a7i »
3zze »
Chlorine in PDB 3zze: Crystal Structure of C-Met Kinase Domain in Complex with N'-((3Z)-4- Chloro-7-Methyl-2-Oxo-1,2-Dihydro-3H-Indol-3-Ylidene)-2-(4- Hydroxyphenyl)Propanohydrazide
Enzymatic activity of Crystal Structure of C-Met Kinase Domain in Complex with N'-((3Z)-4- Chloro-7-Methyl-2-Oxo-1,2-Dihydro-3H-Indol-3-Ylidene)-2-(4- Hydroxyphenyl)Propanohydrazide
All present enzymatic activity of Crystal Structure of C-Met Kinase Domain in Complex with N'-((3Z)-4- Chloro-7-Methyl-2-Oxo-1,2-Dihydro-3H-Indol-3-Ylidene)-2-(4- Hydroxyphenyl)Propanohydrazide:
2.7.10.1;
2.7.10.1;
Protein crystallography data
The structure of Crystal Structure of C-Met Kinase Domain in Complex with N'-((3Z)-4- Chloro-7-Methyl-2-Oxo-1,2-Dihydro-3H-Indol-3-Ylidene)-2-(4- Hydroxyphenyl)Propanohydrazide, PDB code: 3zze
was solved by
M.Mctigue,
Y.Deng,
K.Ryan,
J.J.Cui,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 47.19 / 1.87 |
| Space group | P 21 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 76.864, 94.373, 45.766, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 19.6 / 22.6 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of C-Met Kinase Domain in Complex with N'-((3Z)-4- Chloro-7-Methyl-2-Oxo-1,2-Dihydro-3H-Indol-3-Ylidene)-2-(4- Hydroxyphenyl)Propanohydrazide
(pdb code 3zze). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Crystal Structure of C-Met Kinase Domain in Complex with N'-((3Z)-4- Chloro-7-Methyl-2-Oxo-1,2-Dihydro-3H-Indol-3-Ylidene)-2-(4- Hydroxyphenyl)Propanohydrazide, PDB code: 3zze:
In total only one binding site of Chlorine was determined in the Crystal Structure of C-Met Kinase Domain in Complex with N'-((3Z)-4- Chloro-7-Methyl-2-Oxo-1,2-Dihydro-3H-Indol-3-Ylidene)-2-(4- Hydroxyphenyl)Propanohydrazide, PDB code: 3zze:
Chlorine binding site 1 out of 1 in 3zze
Go back to
Chlorine binding site 1 out
of 1 in the Crystal Structure of C-Met Kinase Domain in Complex with N'-((3Z)-4- Chloro-7-Methyl-2-Oxo-1,2-Dihydro-3H-Indol-3-Ylidene)-2-(4- Hydroxyphenyl)Propanohydrazide
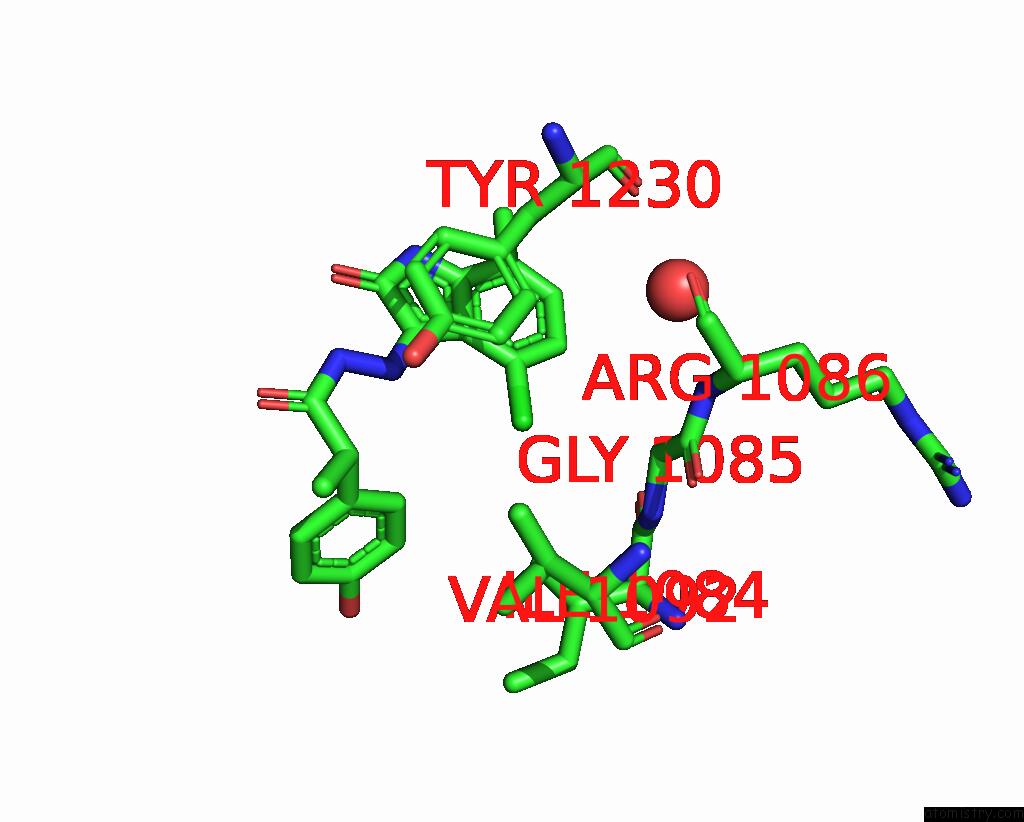
Mono view
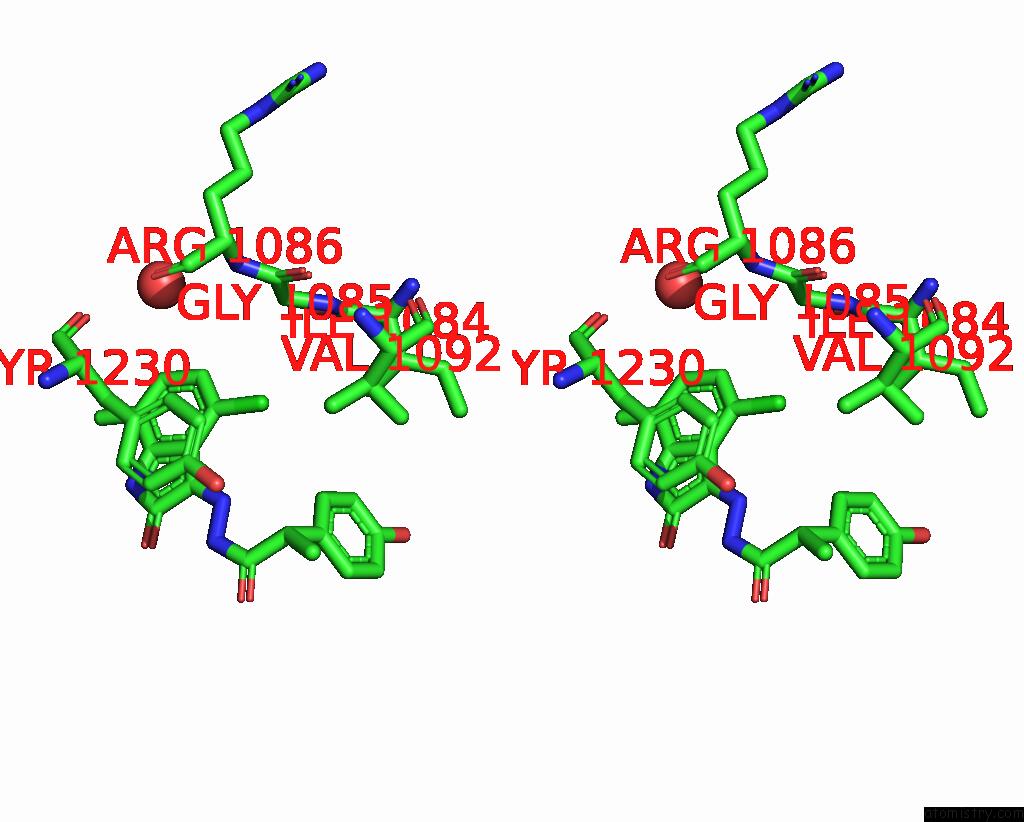
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of C-Met Kinase Domain in Complex with N'-((3Z)-4- Chloro-7-Methyl-2-Oxo-1,2-Dihydro-3H-Indol-3-Ylidene)-2-(4- Hydroxyphenyl)Propanohydrazide within 5.0Å range:
|
Reference:
J.J.Cui,
M.Mctigue,
M.Nambu,
M.Tran-Dube,
M.Pairish,
H.Shen,
L.Jia,
H.Cheng,
J.Hoffman,
P.Le,
M.Jalaie,
G.H.Goetz,
K.Ryan,
N.Grodsky,
Y.Deng,
M.Parker,
S.Timofeevski,
B.W.Murray,
S.Yamazaki,
S.Aguirre,
Q.Li,
H.Zou,
J.Christensen.
Discovery of A Novel Class of Exquisitely Selective Mesenchymal-Epithelial Transition Factor (C-Met) Protein Kinase Inhibitors and Identification of the Clinical Candidate 2-(4-(1-(Quinolin-6-Ylmethyl)-1H-[1,2, 3]Triazolo[4,5-B]Pyrazin-6-Yl)-1H-Pyrazol-1-Yl)Ethanol (Pf-04217903) For the Treatment of Cancer. J.Med.Chem. V. 55 8091 2012.
ISSN: ISSN 0022-2623
PubMed: 22924734
DOI: 10.1021/JM300967G
Page generated: Fri Jul 11 12:35:39 2025
ISSN: ISSN 0022-2623
PubMed: 22924734
DOI: 10.1021/JM300967G
Last articles
Cl in 4MDKCl in 4MCJ
Cl in 4MDQ
Cl in 4MDN
Cl in 4MCS
Cl in 4MCR
Cl in 4MCP
Cl in 4MCQ
Cl in 4MC9
Cl in 4MBZ