Chlorine »
PDB 4ibf-4iij »
4ihu »
Chlorine in PDB 4ihu: Reduced Form of Disulfide Bond Oxdioreductase (Dsbg) From Mycobacterium Tuberculosis
Protein crystallography data
The structure of Reduced Form of Disulfide Bond Oxdioreductase (Dsbg) From Mycobacterium Tuberculosis, PDB code: 4ihu
was solved by
C.A.Harmston,
C.W.Goulding,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 44.70 / 1.90 |
| Space group | P 21 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 71.030, 76.710, 86.960, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.1 / 23.1 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Reduced Form of Disulfide Bond Oxdioreductase (Dsbg) From Mycobacterium Tuberculosis
(pdb code 4ihu). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Reduced Form of Disulfide Bond Oxdioreductase (Dsbg) From Mycobacterium Tuberculosis, PDB code: 4ihu:
In total only one binding site of Chlorine was determined in the Reduced Form of Disulfide Bond Oxdioreductase (Dsbg) From Mycobacterium Tuberculosis, PDB code: 4ihu:
Chlorine binding site 1 out of 1 in 4ihu
Go back to
Chlorine binding site 1 out
of 1 in the Reduced Form of Disulfide Bond Oxdioreductase (Dsbg) From Mycobacterium Tuberculosis
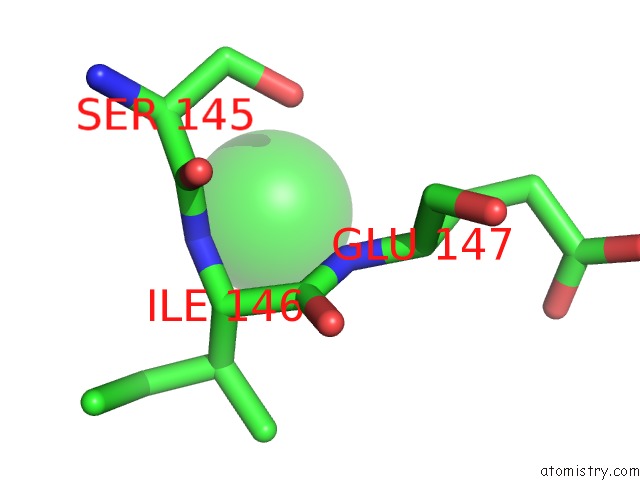
Mono view
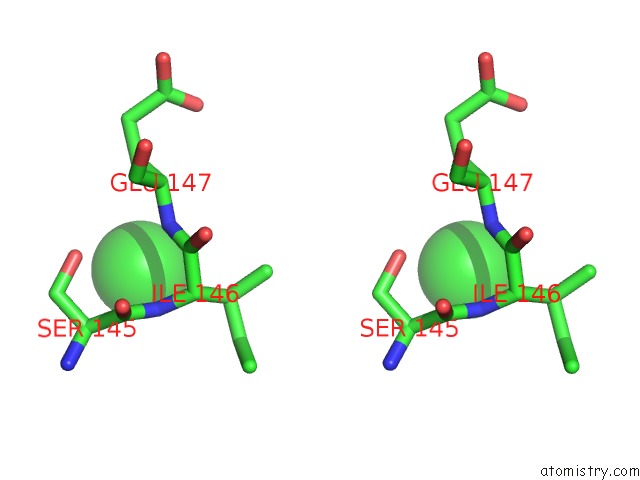
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Reduced Form of Disulfide Bond Oxdioreductase (Dsbg) From Mycobacterium Tuberculosis within 5.0Å range:
|
Reference:
N.Chim,
C.A.Harmston,
D.J.Guzman,
C.W.Goulding.
Structural and Biochemical Characterization of the Essential Dsba-Like Disulfide Bond Forming Protein From Mycobacterium Tuberculosis. Bmc Struct.Biol. V. 13 23 2013.
ISSN: ESSN 1472-6807
PubMed: 24134223
DOI: 10.1186/1472-6807-13-23
Page generated: Fri Jul 11 16:50:58 2025
ISSN: ESSN 1472-6807
PubMed: 24134223
DOI: 10.1186/1472-6807-13-23
Last articles
Cl in 4Y0BCl in 4Y0C
Cl in 4XYM
Cl in 4XZH
Cl in 4XYX
Cl in 4XYL
Cl in 4XYP
Cl in 4XXW
Cl in 4XXX
Cl in 4XXO