Chlorine »
PDB 4jti-4k2a »
4k22 »
Chlorine in PDB 4k22: Structure of the C-Terminal Truncated Form of E.Coli C5-Hydroxylase Ubii Involved in Ubiquinone (Q8) Biosynthesis
Protein crystallography data
The structure of Structure of the C-Terminal Truncated Form of E.Coli C5-Hydroxylase Ubii Involved in Ubiquinone (Q8) Biosynthesis, PDB code: 4k22
was solved by
L.Pecqueur,
M.Lombard,
B.Golinelli-Pimpaneau,
M.Fontecave,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 46.98 / 2.00 |
| Space group | P 21 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 94.730, 124.140, 71.890, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 16.3 / 19.8 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Structure of the C-Terminal Truncated Form of E.Coli C5-Hydroxylase Ubii Involved in Ubiquinone (Q8) Biosynthesis
(pdb code 4k22). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 4 binding sites of Chlorine where determined in the Structure of the C-Terminal Truncated Form of E.Coli C5-Hydroxylase Ubii Involved in Ubiquinone (Q8) Biosynthesis, PDB code: 4k22:
Jump to Chlorine binding site number: 1; 2; 3; 4;
In total 4 binding sites of Chlorine where determined in the Structure of the C-Terminal Truncated Form of E.Coli C5-Hydroxylase Ubii Involved in Ubiquinone (Q8) Biosynthesis, PDB code: 4k22:
Jump to Chlorine binding site number: 1; 2; 3; 4;
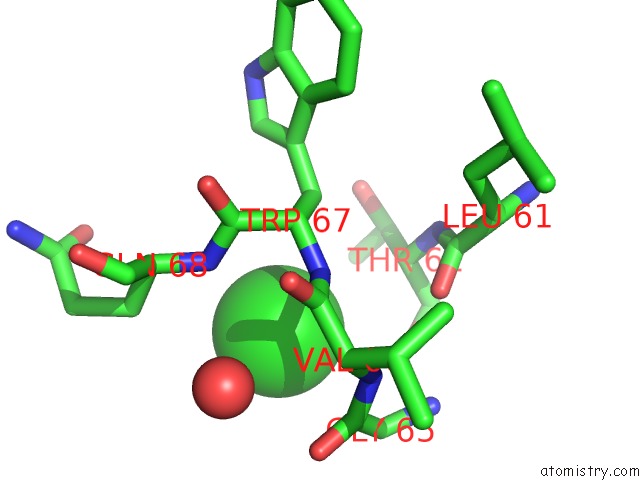
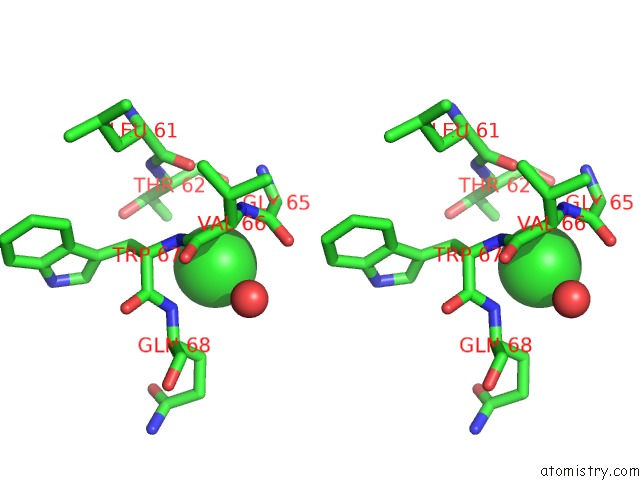
Chlorine binding site 1 out of 4 in 4k22
Go back to
Chlorine binding site 1 out
of 4 in the Structure of the C-Terminal Truncated Form of E.Coli C5-Hydroxylase Ubii Involved in Ubiquinone (Q8) Biosynthesis
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Structure of the C-Terminal Truncated Form of E.Coli C5-Hydroxylase Ubii Involved in Ubiquinone (Q8) Biosynthesis within 5.0Å range:
|
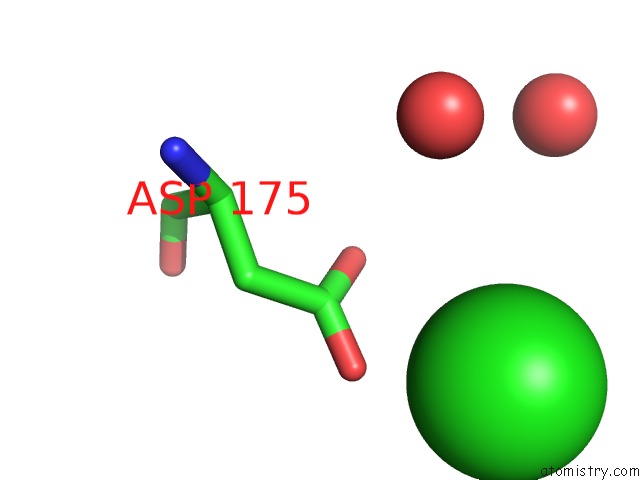
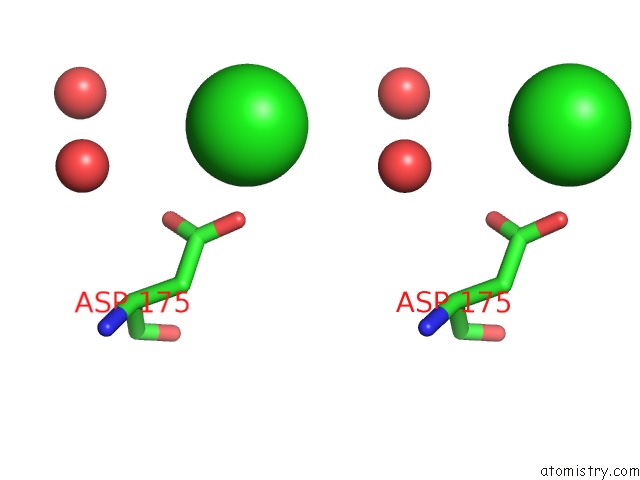
Chlorine binding site 2 out of 4 in 4k22
Go back to
Chlorine binding site 2 out
of 4 in the Structure of the C-Terminal Truncated Form of E.Coli C5-Hydroxylase Ubii Involved in Ubiquinone (Q8) Biosynthesis
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Structure of the C-Terminal Truncated Form of E.Coli C5-Hydroxylase Ubii Involved in Ubiquinone (Q8) Biosynthesis within 5.0Å range:
|
Chlorine binding site 3 out of 4 in 4k22
Go back to
Chlorine binding site 3 out
of 4 in the Structure of the C-Terminal Truncated Form of E.Coli C5-Hydroxylase Ubii Involved in Ubiquinone (Q8) Biosynthesis
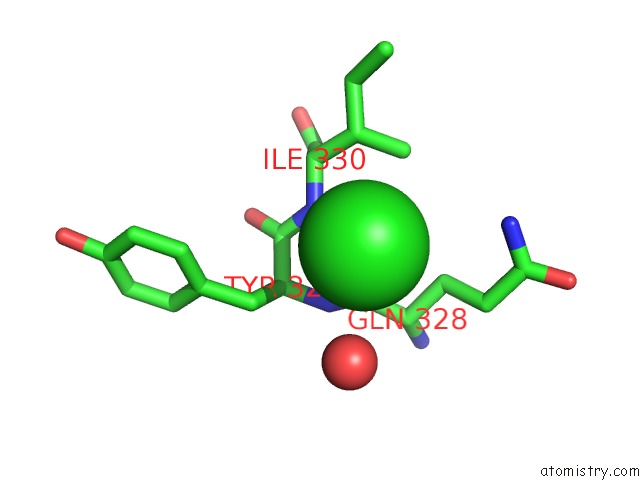
Mono view
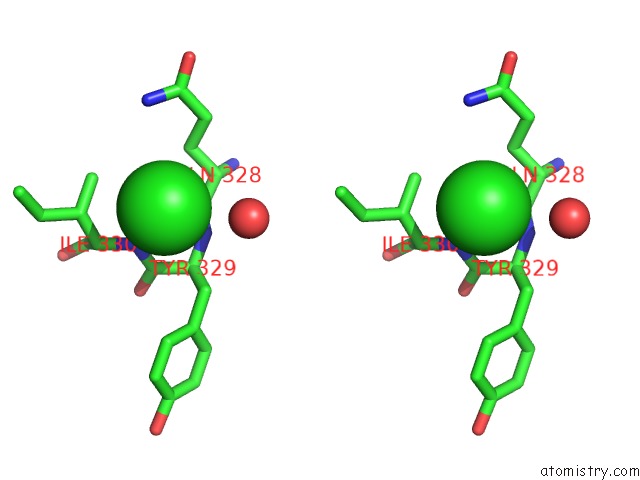
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Structure of the C-Terminal Truncated Form of E.Coli C5-Hydroxylase Ubii Involved in Ubiquinone (Q8) Biosynthesis within 5.0Å range:
|
Chlorine binding site 4 out of 4 in 4k22
Go back to
Chlorine binding site 4 out
of 4 in the Structure of the C-Terminal Truncated Form of E.Coli C5-Hydroxylase Ubii Involved in Ubiquinone (Q8) Biosynthesis
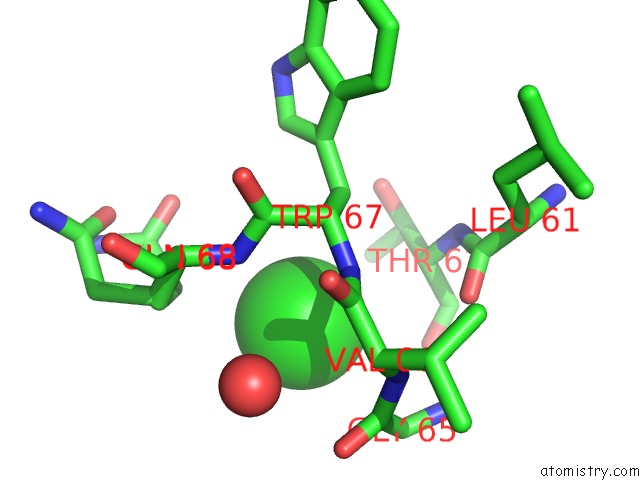
Mono view
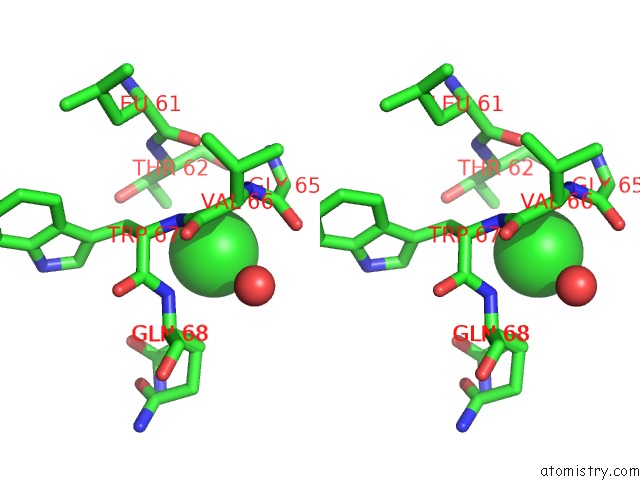
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Structure of the C-Terminal Truncated Form of E.Coli C5-Hydroxylase Ubii Involved in Ubiquinone (Q8) Biosynthesis within 5.0Å range:
|
Reference:
M.H.Chehade,
L.Loiseau,
M.Lombard,
L.Pecqueur,
A.Ismail,
M.Smadja,
B.Golinelli-Pimpaneau,
C.Mellot-Draznieks,
O.Hamelin,
L.Aussel,
S.Kieffer-Jaquinod,
N.Labessan,
F.Barras,
M.Fontecave,
F.Pierrel.
Ubii, A New Gene in Escherichia Coli Coenzyme Q Biosynthesis, Is Involved in Aerobic C5-Hydroxylation. J.Biol.Chem. V. 288 20085 2013.
ISSN: ISSN 0021-9258
PubMed: 23709220
DOI: 10.1074/JBC.M113.480368
Page generated: Fri Jul 11 17:35:51 2025
ISSN: ISSN 0021-9258
PubMed: 23709220
DOI: 10.1074/JBC.M113.480368
Last articles
Cl in 8CHJCl in 8CHN
Cl in 8CHK
Cl in 8CHI
Cl in 8CGS
Cl in 8CGQ
Cl in 8CGP
Cl in 8CGK
Cl in 8CGD
Cl in 8CCT