Chlorine »
PDB 4ogv-4otj »
4op1 »
Chlorine in PDB 4op1: Gkrp Bound to AMG0556 and Sorbitol-6-Phosphate
Protein crystallography data
The structure of Gkrp Bound to AMG0556 and Sorbitol-6-Phosphate, PDB code: 4op1
was solved by
S.R.Jordan,
S.Chmait,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 2.39 |
| Space group | P 65 |
| Cell size a, b, c (Å), α, β, γ (°) | 149.180, 149.180, 132.105, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 20.8 / 26.8 |
Other elements in 4op1:
The structure of Gkrp Bound to AMG0556 and Sorbitol-6-Phosphate also contains other interesting chemical elements:
| Fluorine | (F) | 6 atoms |
| Iodine | (I) | 28 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Gkrp Bound to AMG0556 and Sorbitol-6-Phosphate
(pdb code 4op1). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Gkrp Bound to AMG0556 and Sorbitol-6-Phosphate, PDB code: 4op1:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Gkrp Bound to AMG0556 and Sorbitol-6-Phosphate, PDB code: 4op1:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 4op1
Go back to
Chlorine binding site 1 out
of 2 in the Gkrp Bound to AMG0556 and Sorbitol-6-Phosphate
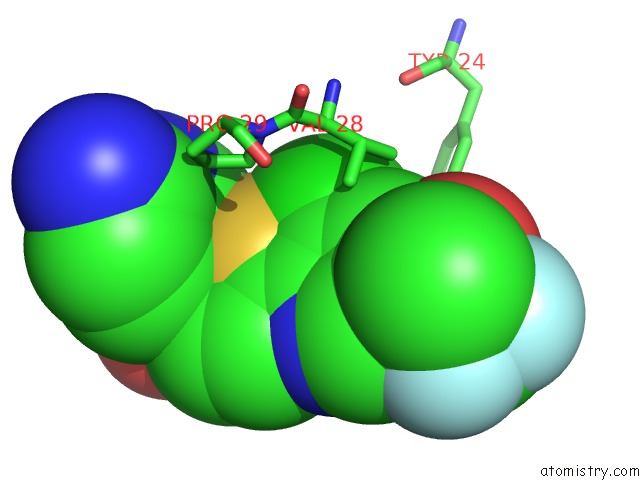
Mono view
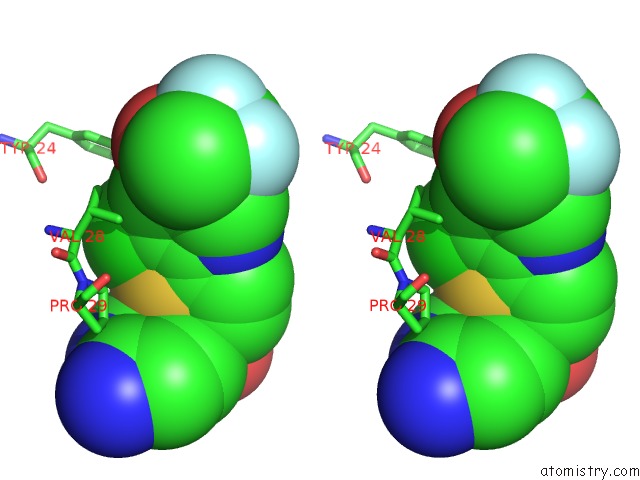
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Gkrp Bound to AMG0556 and Sorbitol-6-Phosphate within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 4op1
Go back to
Chlorine binding site 2 out
of 2 in the Gkrp Bound to AMG0556 and Sorbitol-6-Phosphate
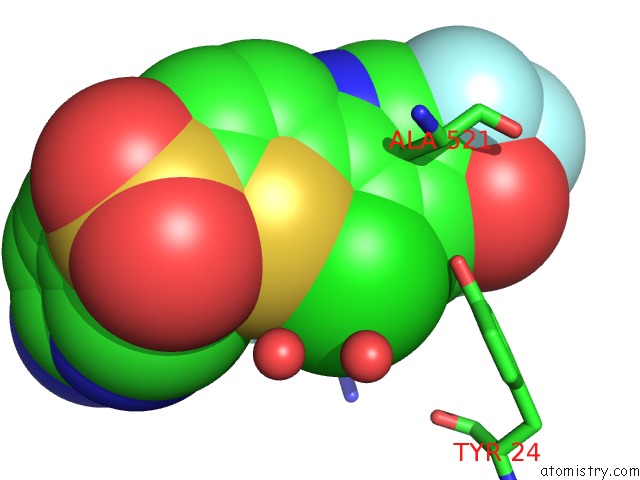
Mono view
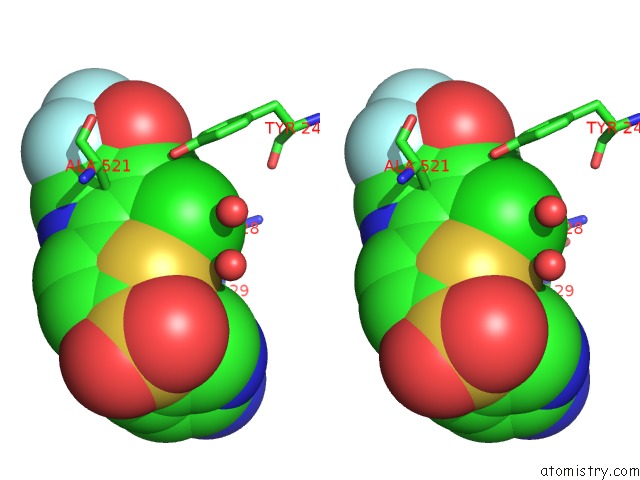
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Gkrp Bound to AMG0556 and Sorbitol-6-Phosphate within 5.0Å range:
|
Reference:
N.Tamayo,
M.H.Norman,
M.D.Bartberger,
Y.Bo,
J.Chen,
R.Cupples,
C.Fotsch,
C.Hale,
F-T Hong,
S.R.Jordan,
L.Liu,
D.J.Lloyd,
C.Moyer,
N.Nishimura,
G.Sivits,
S.Tadesse,
G.Van,
K.C.Yang,
D.J.St. Jean.
Small Molecule Disruptors of the Glucokinase- Glucokinase Regulatory Protein Interaction:5. New Aryl Sulfones Series, Optimization Through Conformational Analysis J.Med.Chem. 2014.
ISSN: ISSN 0022-2623
Page generated: Thu Jul 25 23:32:28 2024
ISSN: ISSN 0022-2623
Last articles
Cl in 2WSMCl in 2WT8
Cl in 2WSL
Cl in 2WSA
Cl in 2WSJ
Cl in 2WS7
Cl in 2WS6
Cl in 2WQ9
Cl in 2WQO
Cl in 2WR6