Chlorine »
PDB 5cu8-5d0x »
5cur »
Chlorine in PDB 5cur: G158E/K44E/R57E/Y49E Bacillus Subtilis Lipase A with 20% [Bmim][Cl]
Enzymatic activity of G158E/K44E/R57E/Y49E Bacillus Subtilis Lipase A with 20% [Bmim][Cl]
All present enzymatic activity of G158E/K44E/R57E/Y49E Bacillus Subtilis Lipase A with 20% [Bmim][Cl]:
3.1.1.3;
3.1.1.3;
Protein crystallography data
The structure of G158E/K44E/R57E/Y49E Bacillus Subtilis Lipase A with 20% [Bmim][Cl], PDB code: 5cur
was solved by
E.M.Nordwald,
J.G.Plaks,
J.R.Snell,
M.C.Sousa,
J.L.Kaar,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 42.50 / 1.30 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 47.721, 56.843, 64.114, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 12.1 / 13.8 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the G158E/K44E/R57E/Y49E Bacillus Subtilis Lipase A with 20% [Bmim][Cl]
(pdb code 5cur). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the G158E/K44E/R57E/Y49E Bacillus Subtilis Lipase A with 20% [Bmim][Cl], PDB code: 5cur:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the G158E/K44E/R57E/Y49E Bacillus Subtilis Lipase A with 20% [Bmim][Cl], PDB code: 5cur:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 5cur
Go back to
Chlorine binding site 1 out
of 2 in the G158E/K44E/R57E/Y49E Bacillus Subtilis Lipase A with 20% [Bmim][Cl]
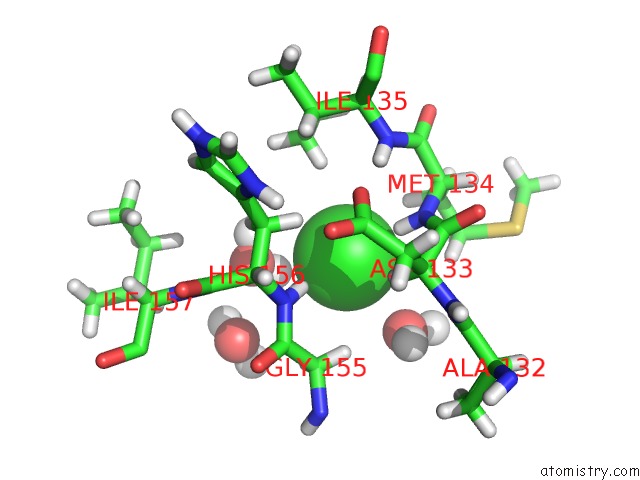
Mono view
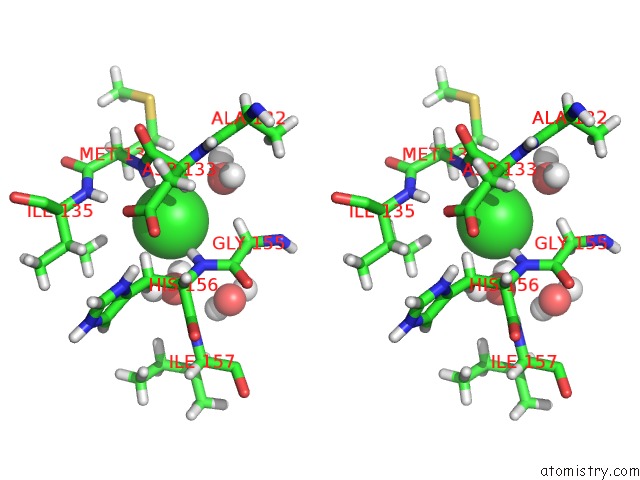
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of G158E/K44E/R57E/Y49E Bacillus Subtilis Lipase A with 20% [Bmim][Cl] within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 5cur
Go back to
Chlorine binding site 2 out
of 2 in the G158E/K44E/R57E/Y49E Bacillus Subtilis Lipase A with 20% [Bmim][Cl]
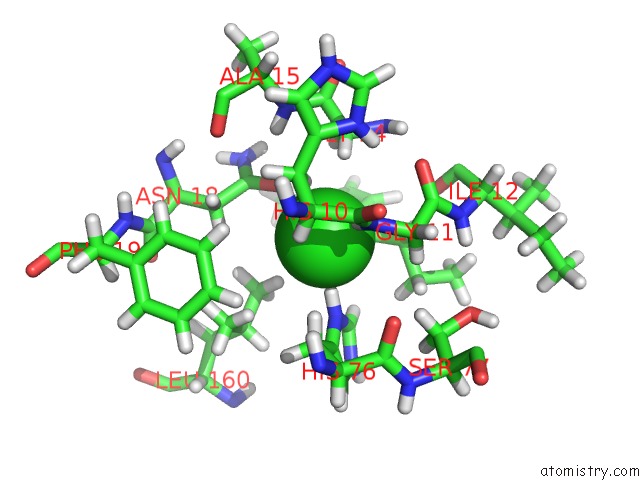
Mono view
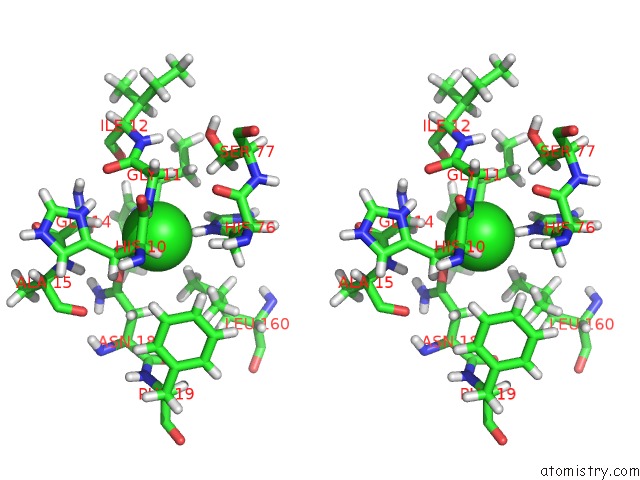
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of G158E/K44E/R57E/Y49E Bacillus Subtilis Lipase A with 20% [Bmim][Cl] within 5.0Å range:
|
Reference:
E.M.Nordwald,
J.G.Plaks,
J.R.Snell,
M.C.Sousa,
J.L.Kaar.
Crystallographic Investigation of Imidazolium Ionic Liquid Effects on Enzyme Structure. Chembiochem V. 16 2456 2015.
ISSN: ESSN 1439-7633
PubMed: 26388426
DOI: 10.1002/CBIC.201500398
Page generated: Fri Jul 26 06:19:02 2024
ISSN: ESSN 1439-7633
PubMed: 26388426
DOI: 10.1002/CBIC.201500398
Last articles
Cl in 2VQ7Cl in 2VQ9
Cl in 2VQ5
Cl in 2VQ8
Cl in 2VQ2
Cl in 2VPM
Cl in 2VPS
Cl in 2VOT
Cl in 2VPQ
Cl in 2VP1