Chlorine »
PDB 5ibp-5ime »
5iez »
Chlorine in PDB 5iez: Discovery of Potent Myeloid Cell Leukemia-1 (Mcl-1) Inhibitors Using Structure-Based Design
Protein crystallography data
The structure of Discovery of Potent Myeloid Cell Leukemia-1 (Mcl-1) Inhibitors Using Structure-Based Design, PDB code: 5iez
was solved by
B.Zhao,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.84 / 2.60 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 39.914, 136.046, 63.499, 90.00, 101.62, 90.00 |
| R / Rfree (%) | 19.4 / 27.7 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Discovery of Potent Myeloid Cell Leukemia-1 (Mcl-1) Inhibitors Using Structure-Based Design
(pdb code 5iez). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 8 binding sites of Chlorine where determined in the Discovery of Potent Myeloid Cell Leukemia-1 (Mcl-1) Inhibitors Using Structure-Based Design, PDB code: 5iez:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
In total 8 binding sites of Chlorine where determined in the Discovery of Potent Myeloid Cell Leukemia-1 (Mcl-1) Inhibitors Using Structure-Based Design, PDB code: 5iez:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
Chlorine binding site 1 out of 8 in 5iez
Go back to
Chlorine binding site 1 out
of 8 in the Discovery of Potent Myeloid Cell Leukemia-1 (Mcl-1) Inhibitors Using Structure-Based Design
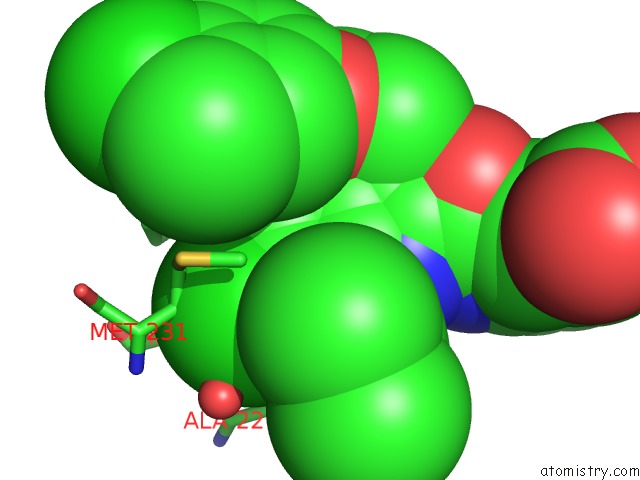
Mono view
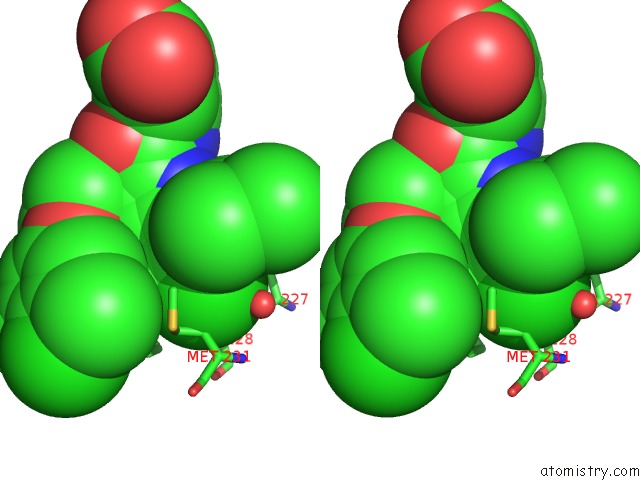
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Discovery of Potent Myeloid Cell Leukemia-1 (Mcl-1) Inhibitors Using Structure-Based Design within 5.0Å range:
|
Chlorine binding site 2 out of 8 in 5iez
Go back to
Chlorine binding site 2 out
of 8 in the Discovery of Potent Myeloid Cell Leukemia-1 (Mcl-1) Inhibitors Using Structure-Based Design
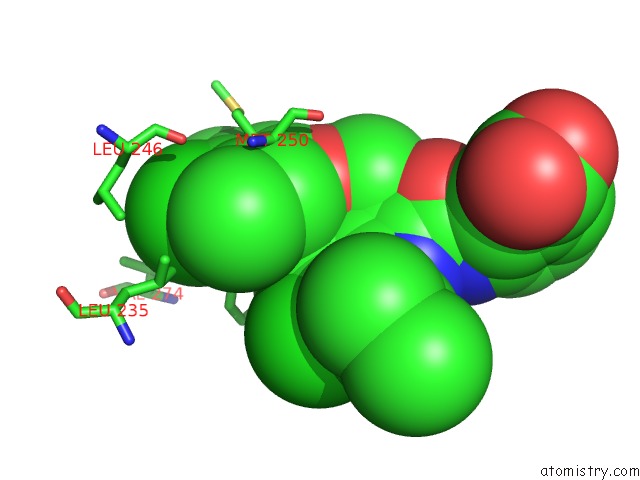
Mono view
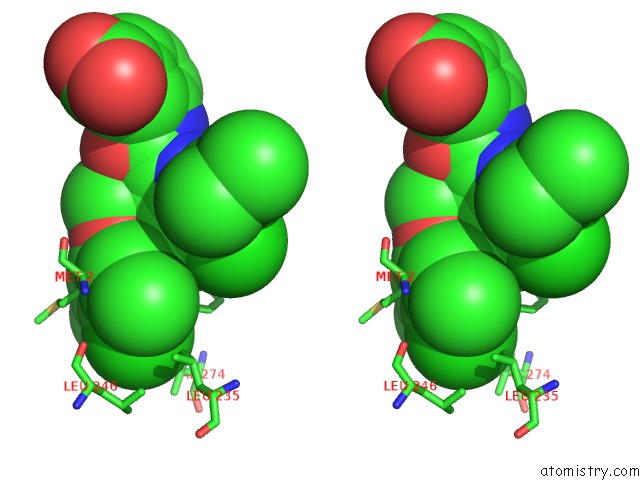
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Discovery of Potent Myeloid Cell Leukemia-1 (Mcl-1) Inhibitors Using Structure-Based Design within 5.0Å range:
|
Chlorine binding site 3 out of 8 in 5iez
Go back to
Chlorine binding site 3 out
of 8 in the Discovery of Potent Myeloid Cell Leukemia-1 (Mcl-1) Inhibitors Using Structure-Based Design
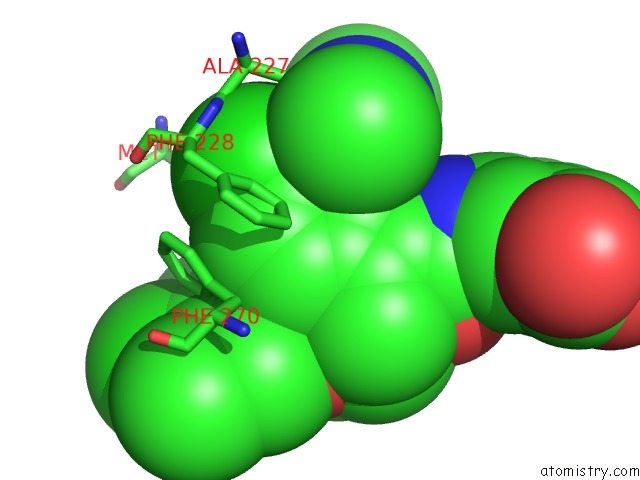
Mono view
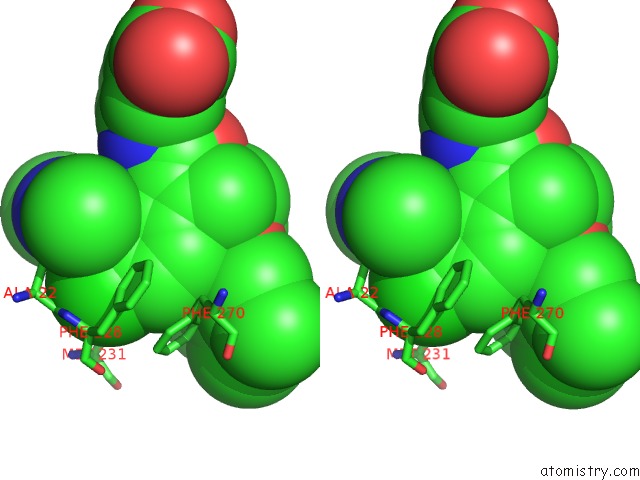
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Discovery of Potent Myeloid Cell Leukemia-1 (Mcl-1) Inhibitors Using Structure-Based Design within 5.0Å range:
|
Chlorine binding site 4 out of 8 in 5iez
Go back to
Chlorine binding site 4 out
of 8 in the Discovery of Potent Myeloid Cell Leukemia-1 (Mcl-1) Inhibitors Using Structure-Based Design
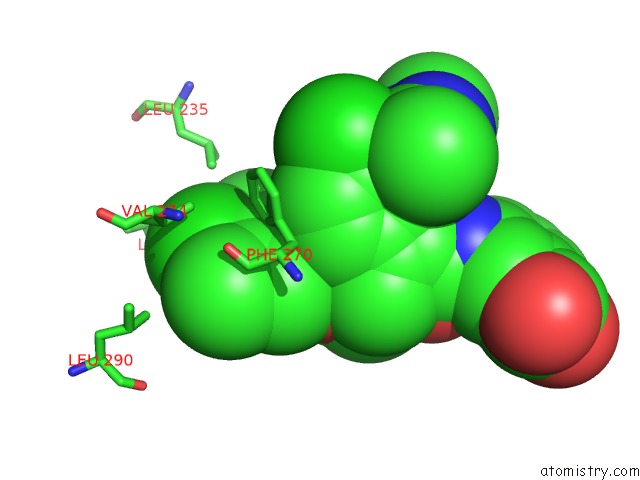
Mono view
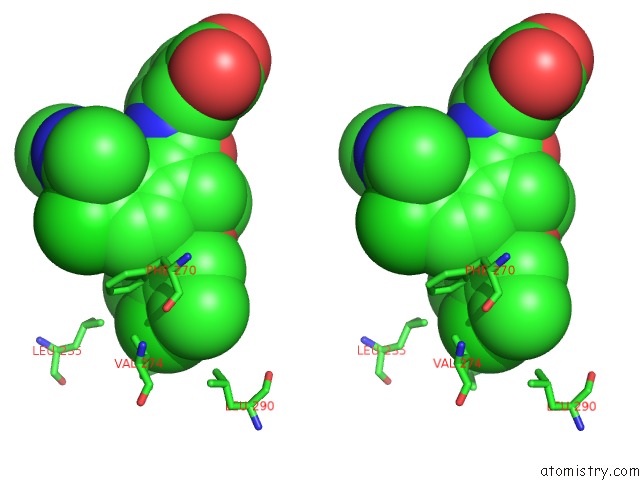
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Discovery of Potent Myeloid Cell Leukemia-1 (Mcl-1) Inhibitors Using Structure-Based Design within 5.0Å range:
|
Chlorine binding site 5 out of 8 in 5iez
Go back to
Chlorine binding site 5 out
of 8 in the Discovery of Potent Myeloid Cell Leukemia-1 (Mcl-1) Inhibitors Using Structure-Based Design
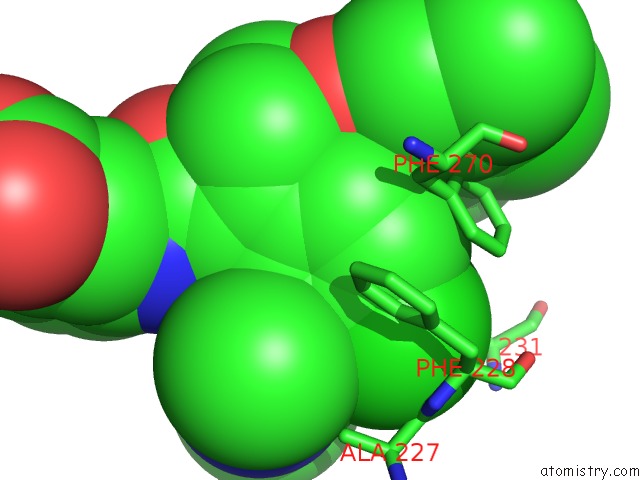
Mono view
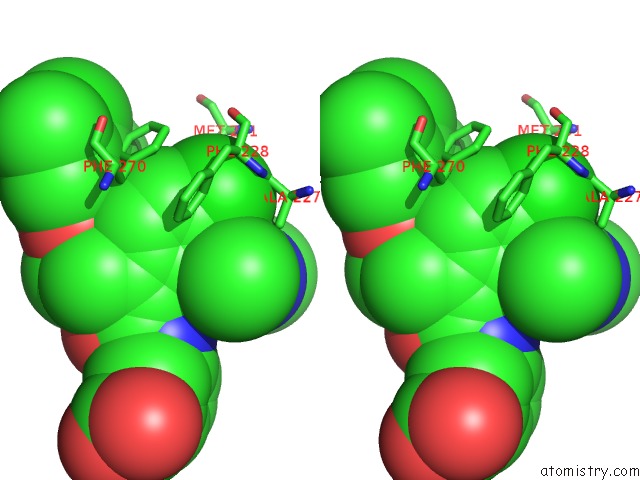
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Discovery of Potent Myeloid Cell Leukemia-1 (Mcl-1) Inhibitors Using Structure-Based Design within 5.0Å range:
|
Chlorine binding site 6 out of 8 in 5iez
Go back to
Chlorine binding site 6 out
of 8 in the Discovery of Potent Myeloid Cell Leukemia-1 (Mcl-1) Inhibitors Using Structure-Based Design
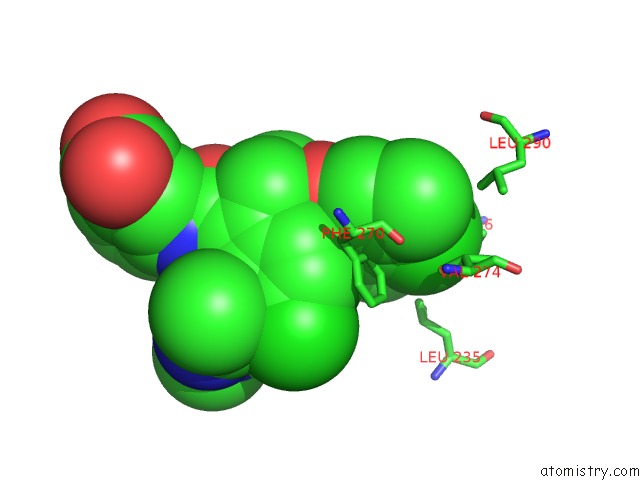
Mono view
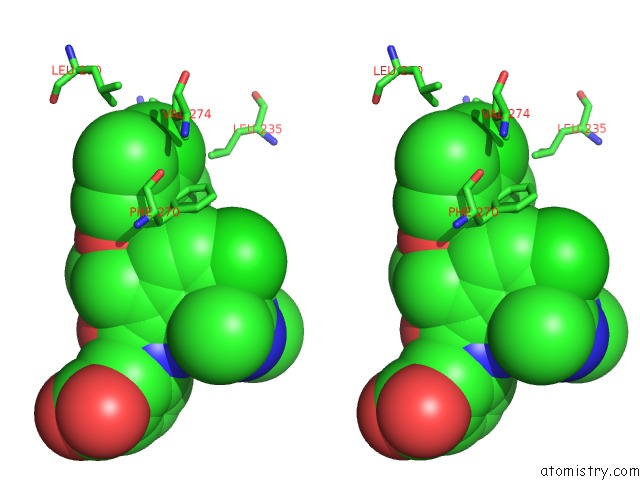
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of Discovery of Potent Myeloid Cell Leukemia-1 (Mcl-1) Inhibitors Using Structure-Based Design within 5.0Å range:
|
Chlorine binding site 7 out of 8 in 5iez
Go back to
Chlorine binding site 7 out
of 8 in the Discovery of Potent Myeloid Cell Leukemia-1 (Mcl-1) Inhibitors Using Structure-Based Design
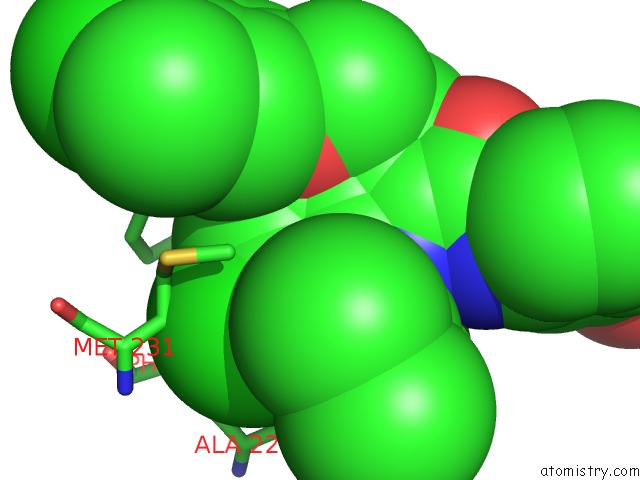
Mono view
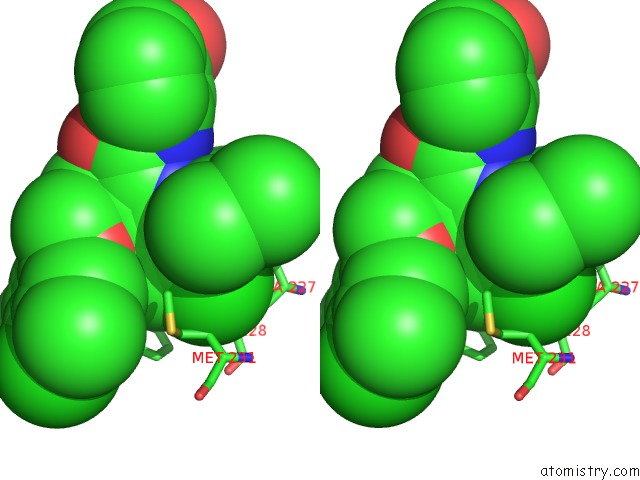
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 7 of Discovery of Potent Myeloid Cell Leukemia-1 (Mcl-1) Inhibitors Using Structure-Based Design within 5.0Å range:
|
Chlorine binding site 8 out of 8 in 5iez
Go back to
Chlorine binding site 8 out
of 8 in the Discovery of Potent Myeloid Cell Leukemia-1 (Mcl-1) Inhibitors Using Structure-Based Design
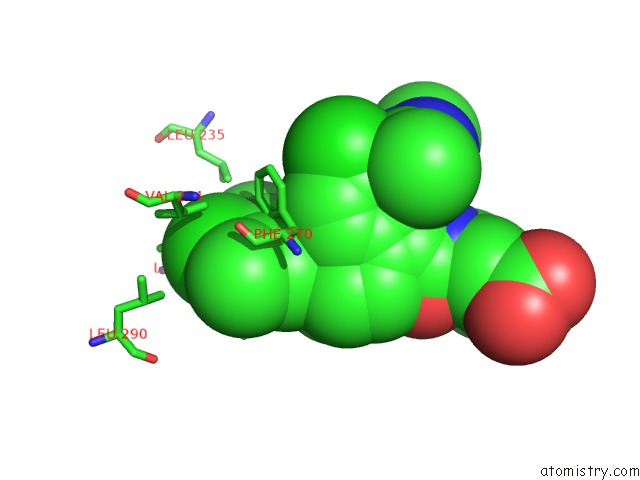
Mono view
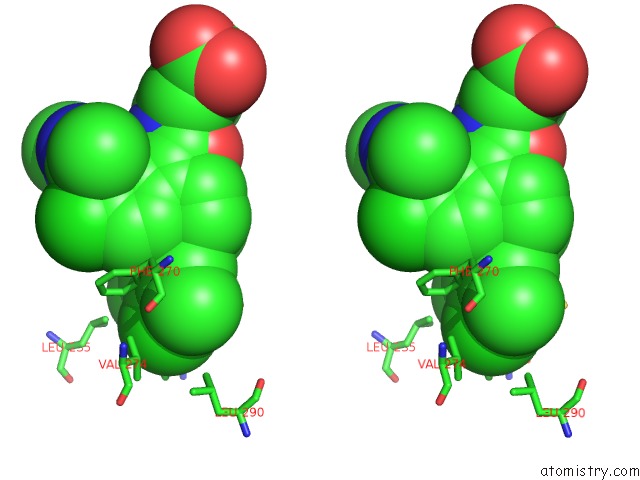
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 8 of Discovery of Potent Myeloid Cell Leukemia-1 (Mcl-1) Inhibitors Using Structure-Based Design within 5.0Å range:
|
Reference:
T.Lee,
Z.Bian,
B.Zhao,
L.J.Hogdal,
J.L.Sensintaffar,
C.M.Goodwin,
J.Belmar,
S.Shaw,
J.C.Tarr,
N.Veerasamy,
S.M.Matulis,
B.Koss,
M.A.Fischer,
A.L.Arnold,
D.V.Camper,
C.F.Browning,
O.W.Rossanese,
A.Budhraja,
J.Opferman,
L.H.Boise,
M.R.Savona,
A.Letai,
E.T.Olejniczak,
S.W.Fesik.
Discovery and Biological Characterization of Potent Myeloid Cell Leukemia-1 Inhibitors. Febs Lett. V. 591 240 2017.
ISSN: ISSN 1873-3468
PubMed: 27878989
DOI: 10.1002/1873-3468.12497
Page generated: Sat Jul 12 03:04:40 2025
ISSN: ISSN 1873-3468
PubMed: 27878989
DOI: 10.1002/1873-3468.12497
Last articles
Cl in 5SKFCl in 5SK8
Cl in 5SK1
Cl in 5SJN
Cl in 5SJK
Cl in 5SJT
Cl in 5SJQ
Cl in 5SJS
Cl in 5SJR
Cl in 5SJI