Chlorine »
PDB 5m86-5mge »
5mac »
Chlorine in PDB 5mac: Crystal Structure of Decameric Methanococcoides Burtonii Rubisco Complexed with 2-Carboxyarabinitol Bisphosphate
Enzymatic activity of Crystal Structure of Decameric Methanococcoides Burtonii Rubisco Complexed with 2-Carboxyarabinitol Bisphosphate
All present enzymatic activity of Crystal Structure of Decameric Methanococcoides Burtonii Rubisco Complexed with 2-Carboxyarabinitol Bisphosphate:
4.1.1.39;
4.1.1.39;
Protein crystallography data
The structure of Crystal Structure of Decameric Methanococcoides Burtonii Rubisco Complexed with 2-Carboxyarabinitol Bisphosphate, PDB code: 5mac
was solved by
L.H.Gunn,
K.Valegard,
I.Andersson,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.37 / 2.60 |
| Space group | P 3 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 273.760, 273.760, 96.740, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 18.9 / 22.5 |
Other elements in 5mac:
The structure of Crystal Structure of Decameric Methanococcoides Burtonii Rubisco Complexed with 2-Carboxyarabinitol Bisphosphate also contains other interesting chemical elements:
| Magnesium | (Mg) | 10 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of Decameric Methanococcoides Burtonii Rubisco Complexed with 2-Carboxyarabinitol Bisphosphate
(pdb code 5mac). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 5 binding sites of Chlorine where determined in the Crystal Structure of Decameric Methanococcoides Burtonii Rubisco Complexed with 2-Carboxyarabinitol Bisphosphate, PDB code: 5mac:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Chlorine where determined in the Crystal Structure of Decameric Methanococcoides Burtonii Rubisco Complexed with 2-Carboxyarabinitol Bisphosphate, PDB code: 5mac:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5;
Chlorine binding site 1 out of 5 in 5mac
Go back to
Chlorine binding site 1 out
of 5 in the Crystal Structure of Decameric Methanococcoides Burtonii Rubisco Complexed with 2-Carboxyarabinitol Bisphosphate
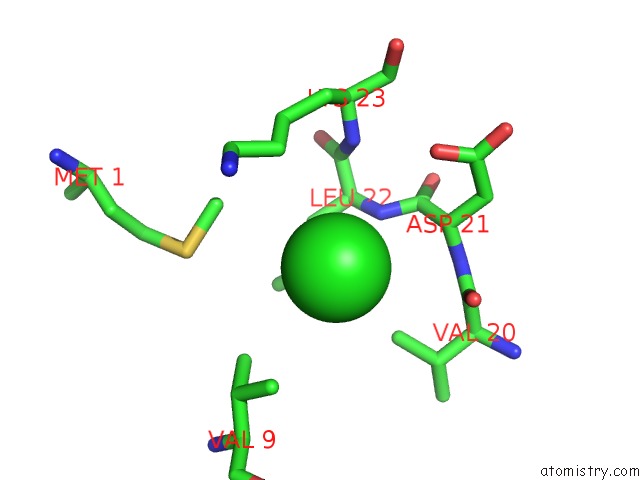
Mono view
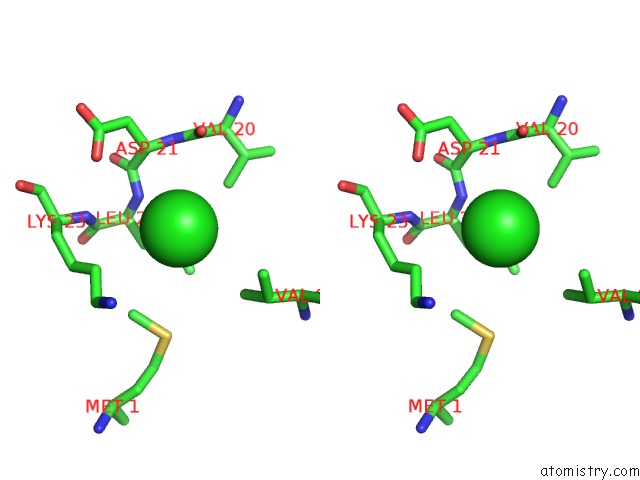
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Decameric Methanococcoides Burtonii Rubisco Complexed with 2-Carboxyarabinitol Bisphosphate within 5.0Å range:
|
Chlorine binding site 2 out of 5 in 5mac
Go back to
Chlorine binding site 2 out
of 5 in the Crystal Structure of Decameric Methanococcoides Burtonii Rubisco Complexed with 2-Carboxyarabinitol Bisphosphate
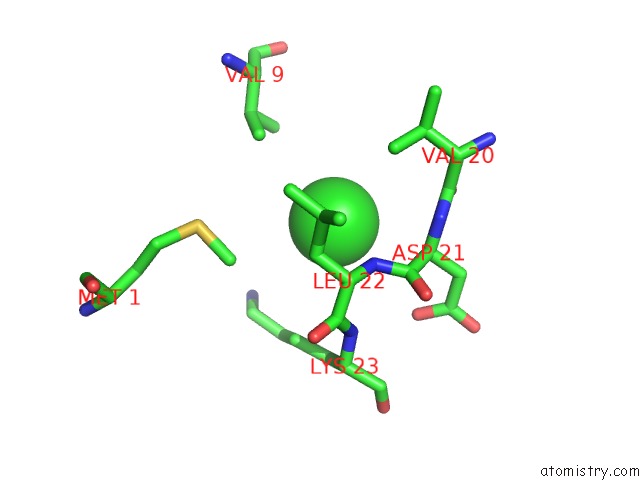
Mono view
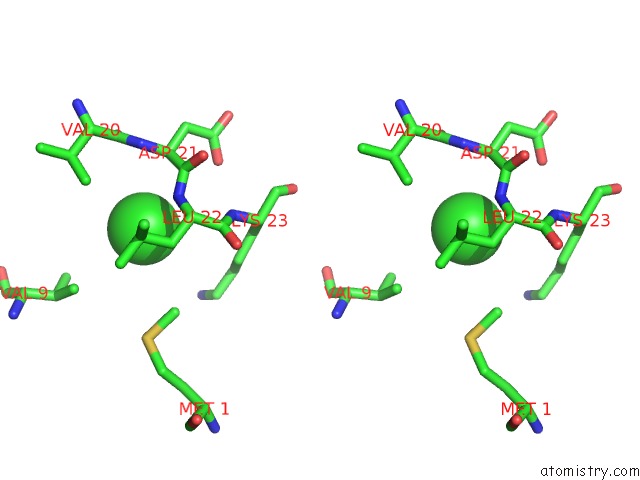
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of Decameric Methanococcoides Burtonii Rubisco Complexed with 2-Carboxyarabinitol Bisphosphate within 5.0Å range:
|
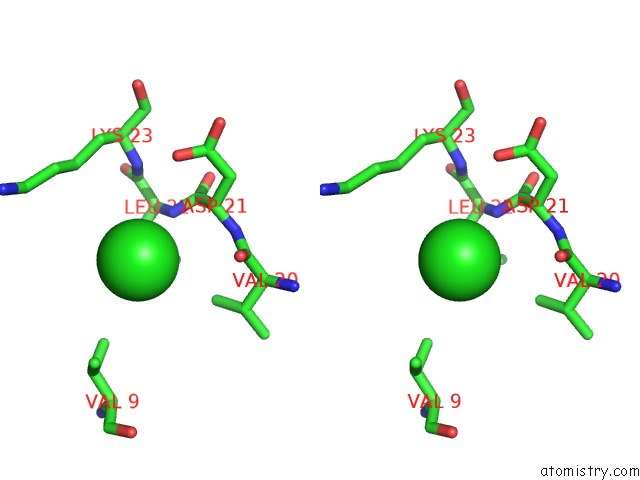
Chlorine binding site 3 out of 5 in 5mac
Go back to
Chlorine binding site 3 out
of 5 in the Crystal Structure of Decameric Methanococcoides Burtonii Rubisco Complexed with 2-Carboxyarabinitol Bisphosphate
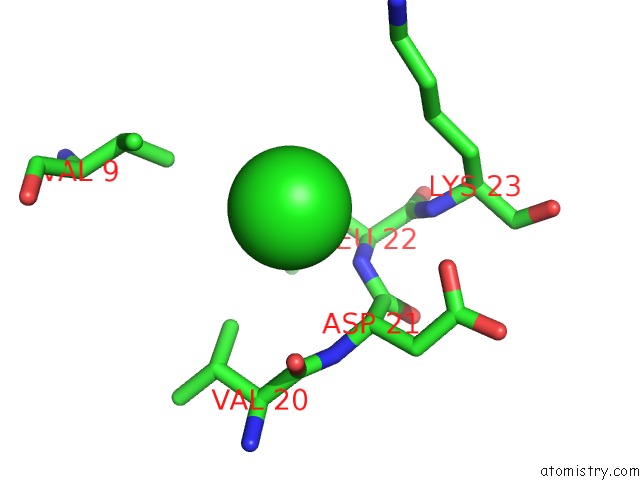
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Crystal Structure of Decameric Methanococcoides Burtonii Rubisco Complexed with 2-Carboxyarabinitol Bisphosphate within 5.0Å range:
|
Chlorine binding site 4 out of 5 in 5mac
Go back to
Chlorine binding site 4 out
of 5 in the Crystal Structure of Decameric Methanococcoides Burtonii Rubisco Complexed with 2-Carboxyarabinitol Bisphosphate
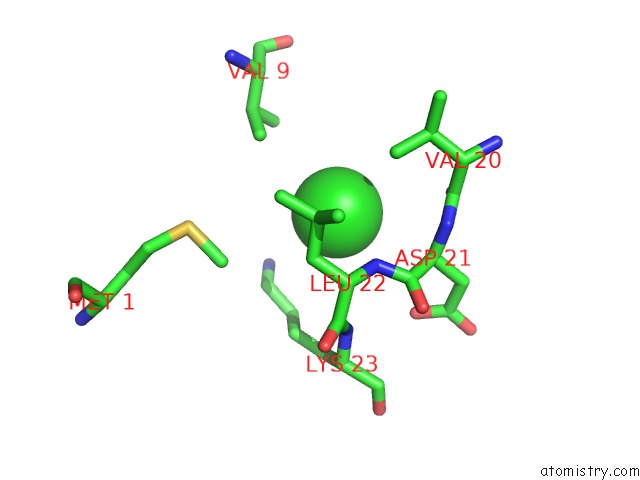
Mono view
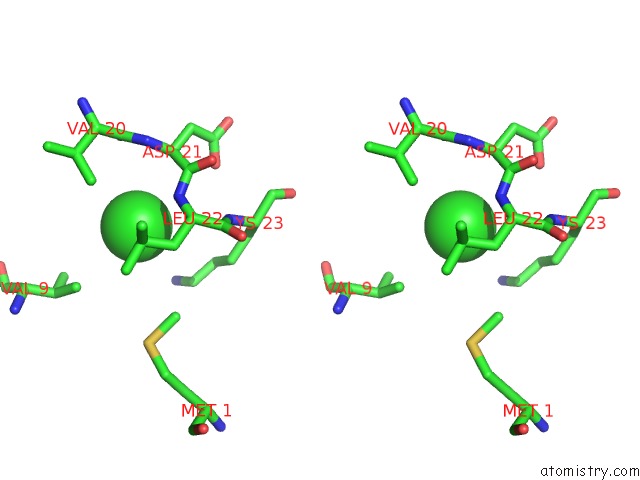
Stereo pair view

Mono view
Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Crystal Structure of Decameric Methanococcoides Burtonii Rubisco Complexed with 2-Carboxyarabinitol Bisphosphate within 5.0Å range:
|
Chlorine binding site 5 out of 5 in 5mac
Go back to
Chlorine binding site 5 out
of 5 in the Crystal Structure of Decameric Methanococcoides Burtonii Rubisco Complexed with 2-Carboxyarabinitol Bisphosphate
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Crystal Structure of Decameric Methanococcoides Burtonii Rubisco Complexed with 2-Carboxyarabinitol Bisphosphate within 5.0Å range:
|
Reference:
L.H.Gunn,
K.Valegard,
I.Andersson.
A Unique Structural Domain in Methanococcoides Burtonii Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase (Rubisco) Acts As A Small Subunit Mimic. J. Biol. Chem. V. 292 6838 2017.
ISSN: ESSN 1083-351X
PubMed: 28154188
DOI: 10.1074/JBC.M116.767145
Page generated: Sat Jul 12 05:23:20 2025
ISSN: ESSN 1083-351X
PubMed: 28154188
DOI: 10.1074/JBC.M116.767145
Last articles
Cl in 5SYXCl in 5SYY
Cl in 5SYM
Cl in 5SYV
Cl in 5SYU
Cl in 5SYL
Cl in 5SYK
Cl in 5SYJ
Cl in 5SYI
Cl in 5SYH