Chlorine »
PDB 5n75-5ncw »
5ncw »
Chlorine in PDB 5ncw: Structure of the Trypsin Induced Serpin-Type Proteinase Inhibitor, Miropin (V367K/K368A Mutant).
Protein crystallography data
The structure of Structure of the Trypsin Induced Serpin-Type Proteinase Inhibitor, Miropin (V367K/K368A Mutant)., PDB code: 5ncw
was solved by
T.Goulas,
M.Ksiazek,
I.Garcia-Ferrer,
D.Mizgalska,
J.Potempa,
X.Gomis-Ruth,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 55.75 / 1.50 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 62.600, 74.280, 84.380, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 16.3 / 18.3 |
Other elements in 5ncw:
The structure of Structure of the Trypsin Induced Serpin-Type Proteinase Inhibitor, Miropin (V367K/K368A Mutant). also contains other interesting chemical elements:
| Potassium | (K) | 1 atom |
| Zinc | (Zn) | 2 atoms |
| Iodine | (I) | 4 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Structure of the Trypsin Induced Serpin-Type Proteinase Inhibitor, Miropin (V367K/K368A Mutant).
(pdb code 5ncw). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 6 binding sites of Chlorine where determined in the Structure of the Trypsin Induced Serpin-Type Proteinase Inhibitor, Miropin (V367K/K368A Mutant)., PDB code: 5ncw:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Chlorine where determined in the Structure of the Trypsin Induced Serpin-Type Proteinase Inhibitor, Miropin (V367K/K368A Mutant)., PDB code: 5ncw:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6;
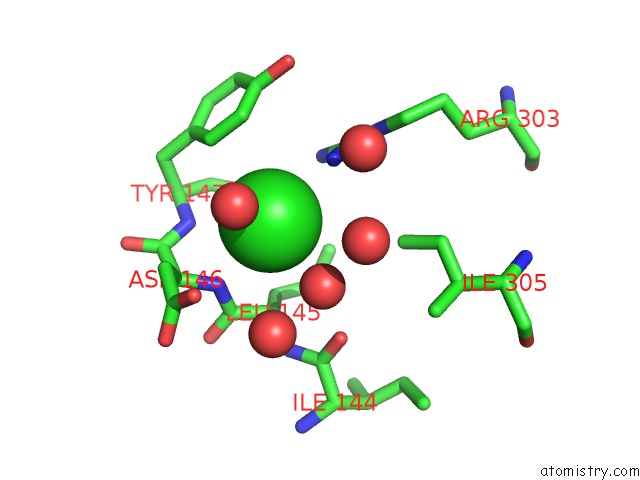
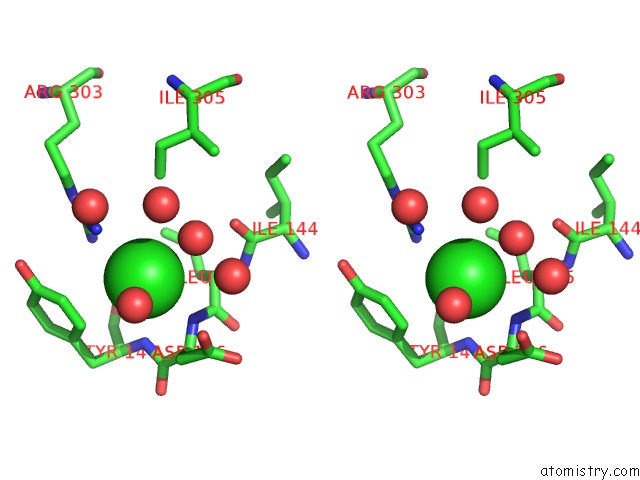
Chlorine binding site 1 out of 6 in 5ncw
Go back to
Chlorine binding site 1 out
of 6 in the Structure of the Trypsin Induced Serpin-Type Proteinase Inhibitor, Miropin (V367K/K368A Mutant).
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Structure of the Trypsin Induced Serpin-Type Proteinase Inhibitor, Miropin (V367K/K368A Mutant). within 5.0Å range:
|
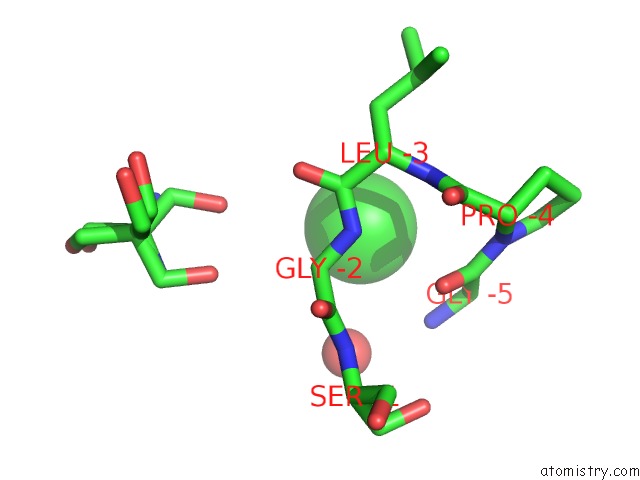
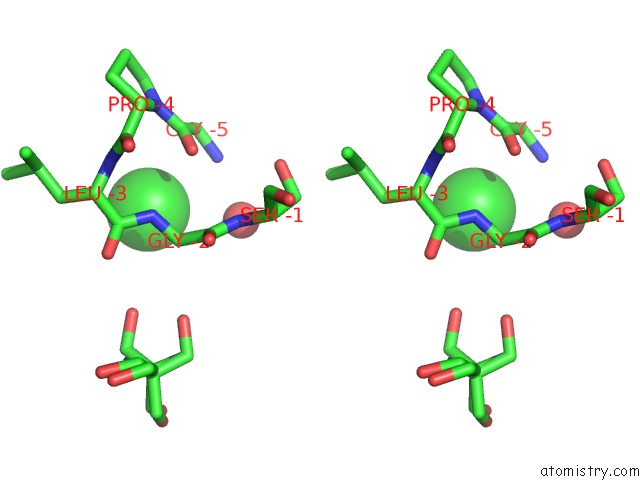
Chlorine binding site 2 out of 6 in 5ncw
Go back to
Chlorine binding site 2 out
of 6 in the Structure of the Trypsin Induced Serpin-Type Proteinase Inhibitor, Miropin (V367K/K368A Mutant).
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Structure of the Trypsin Induced Serpin-Type Proteinase Inhibitor, Miropin (V367K/K368A Mutant). within 5.0Å range:
|
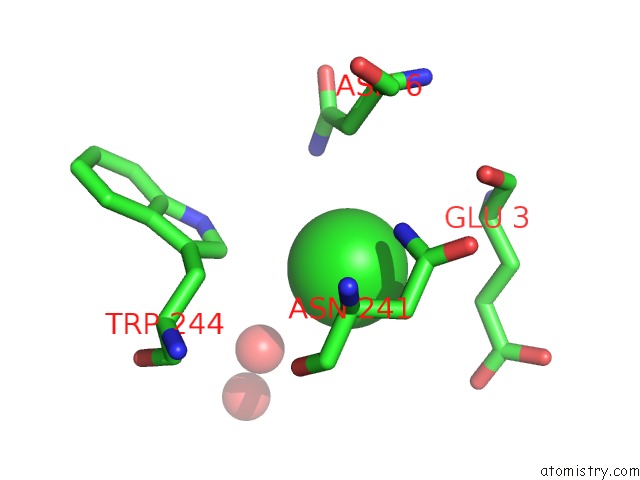
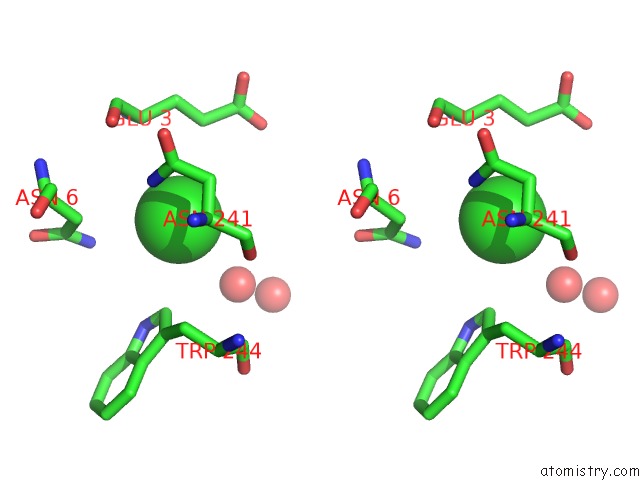
Chlorine binding site 3 out of 6 in 5ncw
Go back to
Chlorine binding site 3 out
of 6 in the Structure of the Trypsin Induced Serpin-Type Proteinase Inhibitor, Miropin (V367K/K368A Mutant).
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Structure of the Trypsin Induced Serpin-Type Proteinase Inhibitor, Miropin (V367K/K368A Mutant). within 5.0Å range:
|
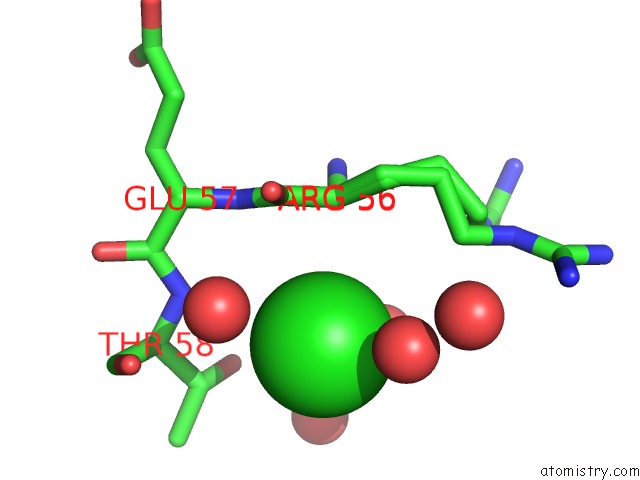
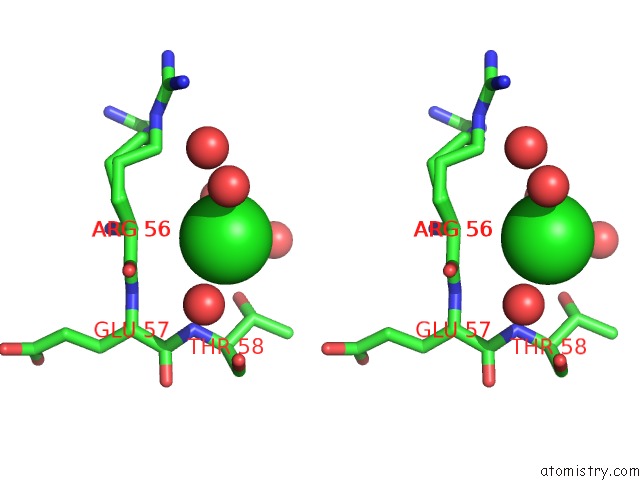
Chlorine binding site 4 out of 6 in 5ncw
Go back to
Chlorine binding site 4 out
of 6 in the Structure of the Trypsin Induced Serpin-Type Proteinase Inhibitor, Miropin (V367K/K368A Mutant).
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Structure of the Trypsin Induced Serpin-Type Proteinase Inhibitor, Miropin (V367K/K368A Mutant). within 5.0Å range:
|
Chlorine binding site 5 out of 6 in 5ncw
Go back to
Chlorine binding site 5 out
of 6 in the Structure of the Trypsin Induced Serpin-Type Proteinase Inhibitor, Miropin (V367K/K368A Mutant).
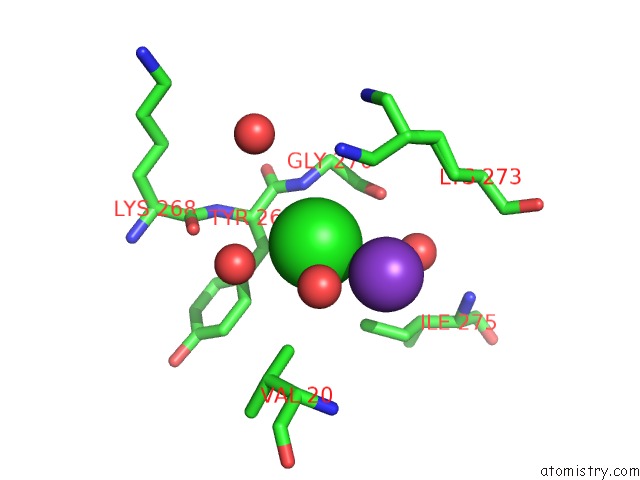
Mono view
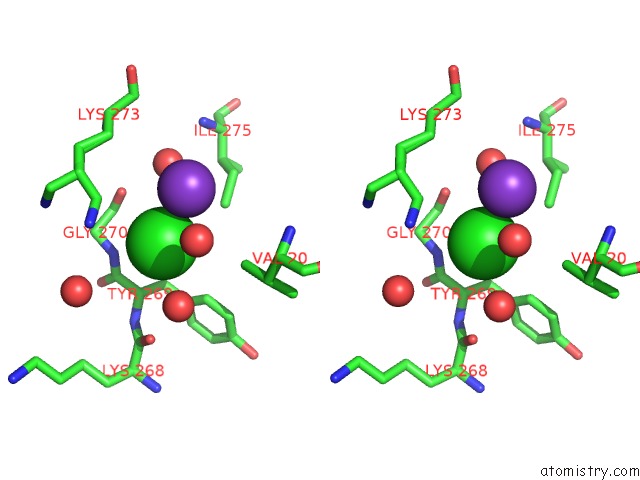
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Structure of the Trypsin Induced Serpin-Type Proteinase Inhibitor, Miropin (V367K/K368A Mutant). within 5.0Å range:
|
Chlorine binding site 6 out of 6 in 5ncw
Go back to
Chlorine binding site 6 out
of 6 in the Structure of the Trypsin Induced Serpin-Type Proteinase Inhibitor, Miropin (V367K/K368A Mutant).
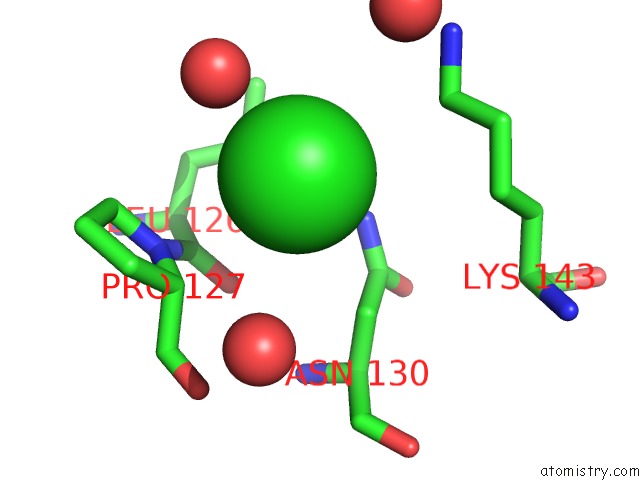
Mono view
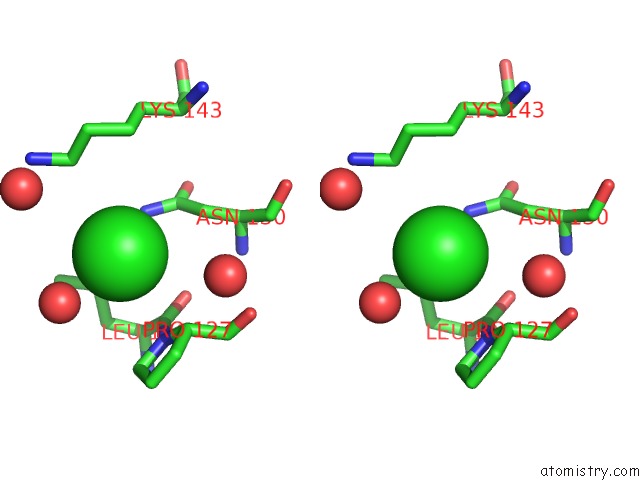
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of Structure of the Trypsin Induced Serpin-Type Proteinase Inhibitor, Miropin (V367K/K368A Mutant). within 5.0Å range:
|
Reference:
T.Goulas,
M.Ksiazek,
I.Garcia-Ferrer,
A.M.Sochaj-Gregorczyk,
I.Waligorska,
M.Wasylewski,
J.Potempa,
F.X.Gomis-Ruth.
A Structure-Derived Snap-Trap Mechanism of A Multispecific Serpin From the Dysbiotic Human Oral Microbiome. J. Biol. Chem. V. 292 10883 2017.
ISSN: ESSN 1083-351X
PubMed: 28512127
DOI: 10.1074/JBC.M117.786533
Page generated: Sat Jul 12 06:03:31 2025
ISSN: ESSN 1083-351X
PubMed: 28512127
DOI: 10.1074/JBC.M117.786533
Last articles
Cl in 5UGHCl in 5UH0
Cl in 5UGR
Cl in 5UFU
Cl in 5UFR
Cl in 5UFQ
Cl in 5UFO
Cl in 5UFE
Cl in 5UEU
Cl in 5UF1