Chlorine »
PDB 5nke-5nry »
5nmu »
Chlorine in PDB 5nmu: Structure of Hexameric Cbs-CP12 Protein From Bloom-Forming Cyanobacteria
Protein crystallography data
The structure of Structure of Hexameric Cbs-CP12 Protein From Bloom-Forming Cyanobacteria, PDB code: 5nmu
was solved by
C.Hackenberg,
J.Hakanpaa,
S.V.Antonyuk,
E.Dittmann,
V.S.Lamzin,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 38.58 / 2.15 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 81.607, 124.021, 118.447, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.8 / 23.4 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Structure of Hexameric Cbs-CP12 Protein From Bloom-Forming Cyanobacteria
(pdb code 5nmu). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 4 binding sites of Chlorine where determined in the Structure of Hexameric Cbs-CP12 Protein From Bloom-Forming Cyanobacteria, PDB code: 5nmu:
Jump to Chlorine binding site number: 1; 2; 3; 4;
In total 4 binding sites of Chlorine where determined in the Structure of Hexameric Cbs-CP12 Protein From Bloom-Forming Cyanobacteria, PDB code: 5nmu:
Jump to Chlorine binding site number: 1; 2; 3; 4;
Chlorine binding site 1 out of 4 in 5nmu
Go back to
Chlorine binding site 1 out
of 4 in the Structure of Hexameric Cbs-CP12 Protein From Bloom-Forming Cyanobacteria
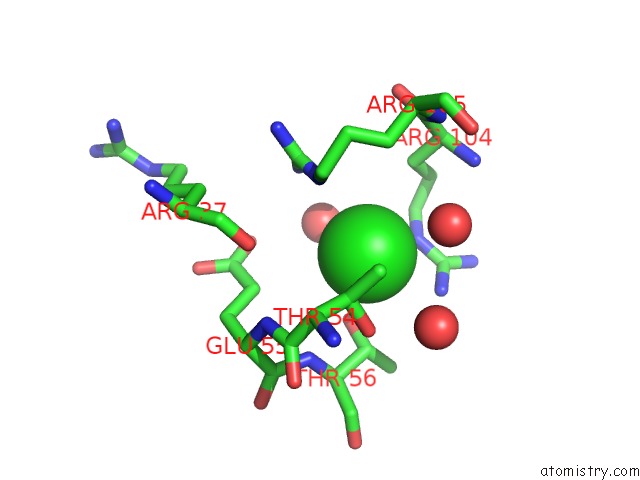
Mono view
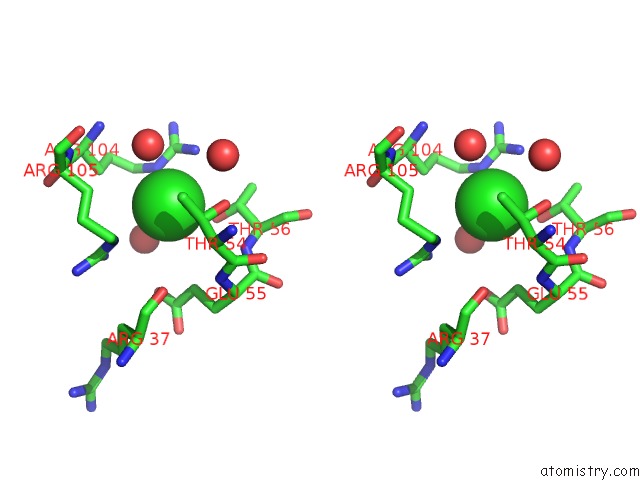
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Structure of Hexameric Cbs-CP12 Protein From Bloom-Forming Cyanobacteria within 5.0Å range:
|
Chlorine binding site 2 out of 4 in 5nmu
Go back to
Chlorine binding site 2 out
of 4 in the Structure of Hexameric Cbs-CP12 Protein From Bloom-Forming Cyanobacteria
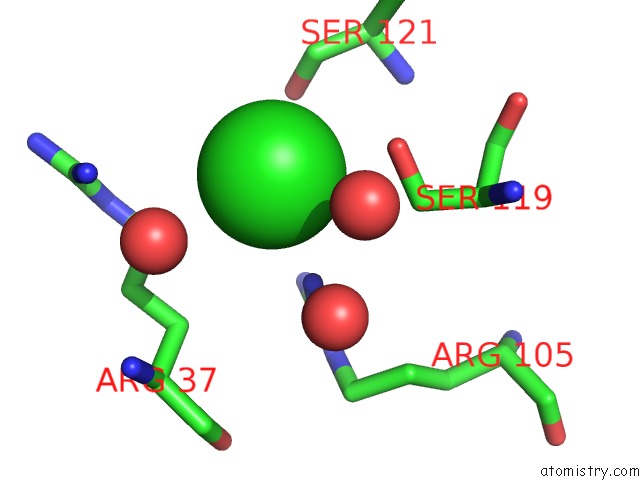
Mono view
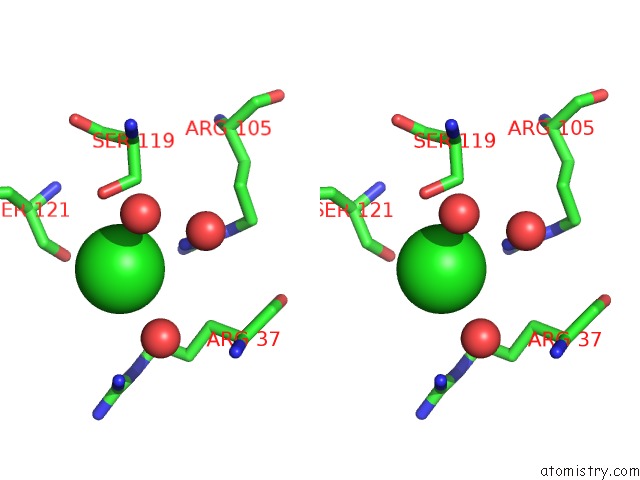
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Structure of Hexameric Cbs-CP12 Protein From Bloom-Forming Cyanobacteria within 5.0Å range:
|
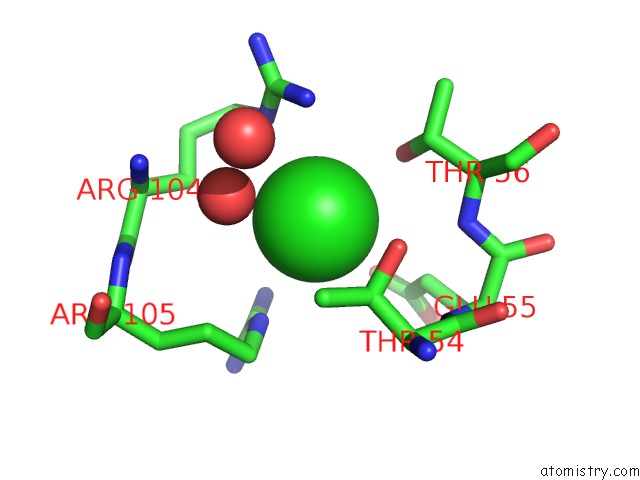
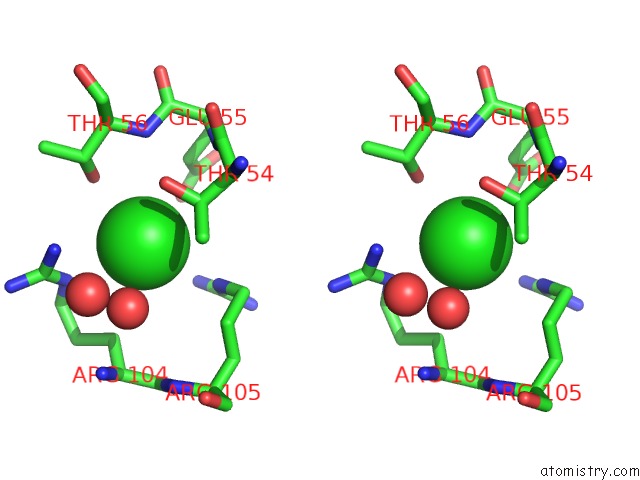
Chlorine binding site 3 out of 4 in 5nmu
Go back to
Chlorine binding site 3 out
of 4 in the Structure of Hexameric Cbs-CP12 Protein From Bloom-Forming Cyanobacteria
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Structure of Hexameric Cbs-CP12 Protein From Bloom-Forming Cyanobacteria within 5.0Å range:
|
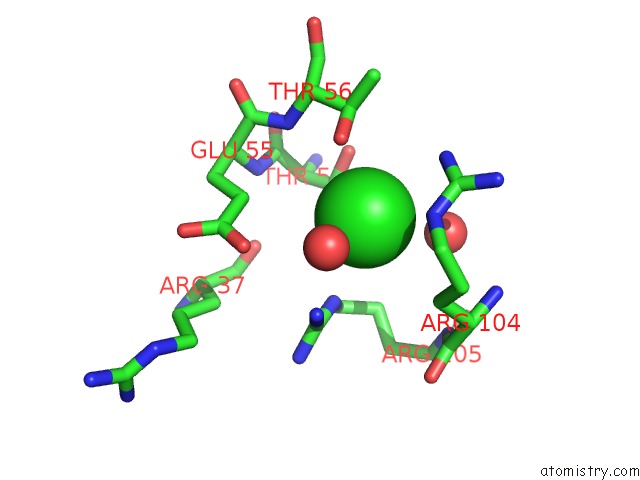
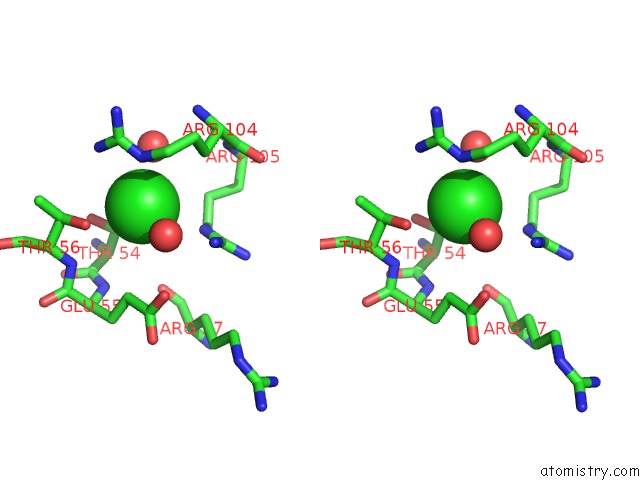
Chlorine binding site 4 out of 4 in 5nmu
Go back to
Chlorine binding site 4 out
of 4 in the Structure of Hexameric Cbs-CP12 Protein From Bloom-Forming Cyanobacteria
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Structure of Hexameric Cbs-CP12 Protein From Bloom-Forming Cyanobacteria within 5.0Å range:
|
Reference:
C.Hackenberg,
J.Hakanpaa,
F.Cai,
S.Antonyuk,
C.Eigner,
S.Meissner,
M.Laitaoja,
J.Janis,
C.A.Kerfeld,
E.Dittmann,
V.S.Lamzin.
Structural and Functional Insights Into the Unique Cbs-CP12 Fusion Protein Family in Cyanobacteria. Proc. Natl. Acad. Sci. V. 115 7141 2018U.S.A..
ISSN: ESSN 1091-6490
PubMed: 29915055
DOI: 10.1073/PNAS.1806668115
Page generated: Sat Jul 12 06:15:02 2025
ISSN: ESSN 1091-6490
PubMed: 29915055
DOI: 10.1073/PNAS.1806668115
Last articles
Cl in 6A8CCl in 6AB2
Cl in 6AB9
Cl in 6A8A
Cl in 6A8B
Cl in 6A94
Cl in 6A7P
Cl in 6A84
Cl in 6A7D
Cl in 6A5Z