Chlorine »
PDB 6cub-6d3h »
6cvy »
Chlorine in PDB 6cvy: Crystal Structure of Hcv NS3/4A Wt Protease in Complex with Aj-21 (Mk- 5172 Linear Analogue)
Protein crystallography data
The structure of Crystal Structure of Hcv NS3/4A Wt Protease in Complex with Aj-21 (Mk- 5172 Linear Analogue), PDB code: 6cvy
was solved by
A.N.Matthew,
C.A.Schiffer,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 27.73 / 1.80 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 55.453, 58.483, 59.729, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 14.7 / 18.4 |
Other elements in 6cvy:
The structure of Crystal Structure of Hcv NS3/4A Wt Protease in Complex with Aj-21 (Mk- 5172 Linear Analogue) also contains other interesting chemical elements:
| Zinc | (Zn) | 1 atom |
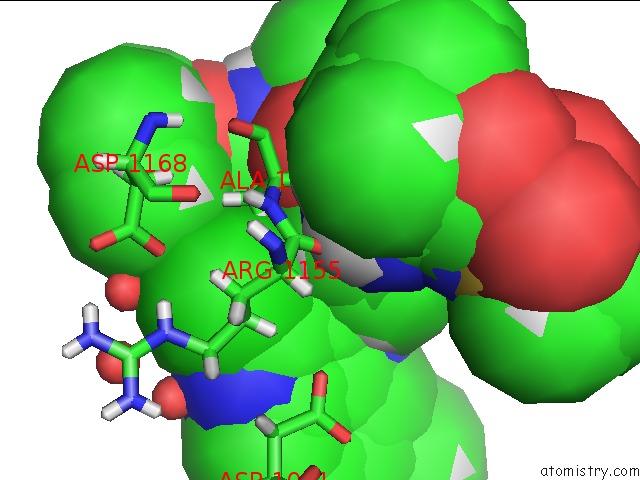
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of Hcv NS3/4A Wt Protease in Complex with Aj-21 (Mk- 5172 Linear Analogue)
(pdb code 6cvy). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Crystal Structure of Hcv NS3/4A Wt Protease in Complex with Aj-21 (Mk- 5172 Linear Analogue), PDB code: 6cvy:
In total only one binding site of Chlorine was determined in the Crystal Structure of Hcv NS3/4A Wt Protease in Complex with Aj-21 (Mk- 5172 Linear Analogue), PDB code: 6cvy:
Chlorine binding site 1 out of 1 in 6cvy
Go back to
Chlorine binding site 1 out
of 1 in the Crystal Structure of Hcv NS3/4A Wt Protease in Complex with Aj-21 (Mk- 5172 Linear Analogue)
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Hcv NS3/4A Wt Protease in Complex with Aj-21 (Mk- 5172 Linear Analogue) within 5.0Å range:
|
Reference:
L.N.Rusere,
A.N.Matthew,
G.J.Lockbaum,
M.Jahangir,
A.Newton,
C.J.Petropoulos,
W.Huang,
N.Kurt Yilmaz,
C.A.Schiffer,
A.Ali.
Quinoxaline-Based Linear Hcv NS3/4A Protease Inhibitors Exhibit Potent Activity Against Drug Resistant Variants. Acs Med Chem Lett V. 9 691 2018.
ISSN: ISSN 1948-5875
PubMed: 30034602
DOI: 10.1021/ACSMEDCHEMLETT.8B00150
Page generated: Sat Jul 27 21:12:37 2024
ISSN: ISSN 1948-5875
PubMed: 30034602
DOI: 10.1021/ACSMEDCHEMLETT.8B00150
Last articles
Cl in 5SQRCl in 5SPQ
Cl in 5SQH
Cl in 5SPO
Cl in 5SPK
Cl in 5SP4
Cl in 5SPJ
Cl in 5SOS
Cl in 5SOK
Cl in 5SNJ