Chlorine »
PDB 6frx-6g1u »
6g13 »
Chlorine in PDB 6g13: C-Terminal Domain of Mers-Cov Nucleocapsid
Protein crystallography data
The structure of C-Terminal Domain of Mers-Cov Nucleocapsid, PDB code: 6g13
was solved by
T.H.V.Nguyen,
F.P.Ferron,
J.Lichiere,
B.Canard,
N.Papageorgiou,
B.Coutard,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 69.33 / 1.97 |
| Space group | P 31 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 120.680, 120.680, 92.646, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 19 / 21.6 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the C-Terminal Domain of Mers-Cov Nucleocapsid
(pdb code 6g13). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the C-Terminal Domain of Mers-Cov Nucleocapsid, PDB code: 6g13:
In total only one binding site of Chlorine was determined in the C-Terminal Domain of Mers-Cov Nucleocapsid, PDB code: 6g13:
Chlorine binding site 1 out of 1 in 6g13
Go back to
Chlorine binding site 1 out
of 1 in the C-Terminal Domain of Mers-Cov Nucleocapsid
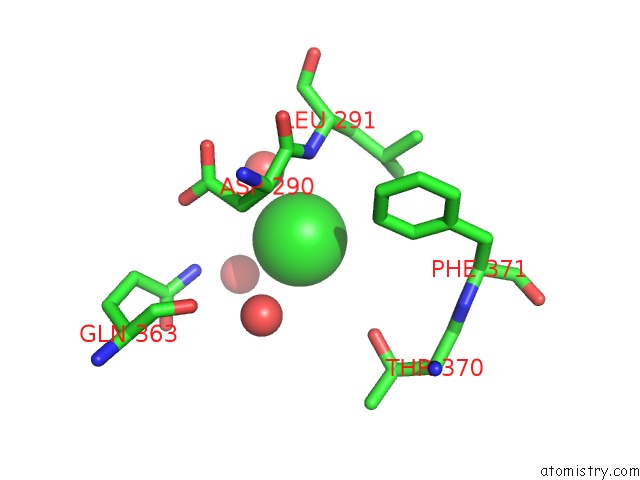
Mono view
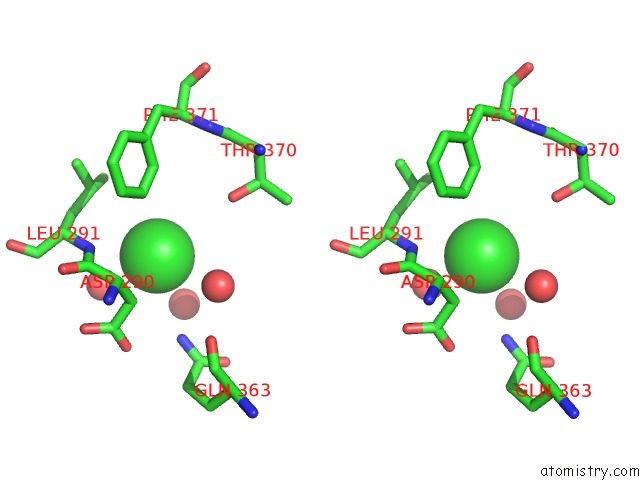
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of C-Terminal Domain of Mers-Cov Nucleocapsid within 5.0Å range:
|
Reference:
T.H.V.Nguyen,
J.Lichiere,
B.Canard,
N.Papageorgiou,
S.Attoumani,
F.Ferron,
B.Coutard.
Structure and Oligomerization State of the C-Terminal Region of the Middle East Respiratory Syndrome Coronavirus Nucleoprotein. Acta Crystallogr D Struct V. 75 8 2019BIOL.
ISSN: ISSN 2059-7983
PubMed: 30644840
DOI: 10.1107/S2059798318014948
Page generated: Sat Jul 12 14:18:17 2025
ISSN: ISSN 2059-7983
PubMed: 30644840
DOI: 10.1107/S2059798318014948
Last articles
Cl in 6PP9Cl in 6PP2
Cl in 6PP1
Cl in 6PP0
Cl in 6PKY
Cl in 6POZ
Cl in 6POY
Cl in 6PLG
Cl in 6POX
Cl in 6POW