Chlorine »
PDB 6hzz-6i6c »
6i29 »
Chlorine in PDB 6i29: X-Ray Structure of the P53-MDM2 Inhibitor NMI801 Bound to HDM2 at 2.1A Resolution
Protein crystallography data
The structure of X-Ray Structure of the P53-MDM2 Inhibitor NMI801 Bound to HDM2 at 2.1A Resolution, PDB code: 6i29
was solved by
J.Kallen,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 19.27 / 2.10 |
| Space group | I 41 3 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 121.883, 121.883, 121.883, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 24.3 / 27.7 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the X-Ray Structure of the P53-MDM2 Inhibitor NMI801 Bound to HDM2 at 2.1A Resolution
(pdb code 6i29). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the X-Ray Structure of the P53-MDM2 Inhibitor NMI801 Bound to HDM2 at 2.1A Resolution, PDB code: 6i29:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the X-Ray Structure of the P53-MDM2 Inhibitor NMI801 Bound to HDM2 at 2.1A Resolution, PDB code: 6i29:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 6i29
Go back to
Chlorine binding site 1 out
of 2 in the X-Ray Structure of the P53-MDM2 Inhibitor NMI801 Bound to HDM2 at 2.1A Resolution
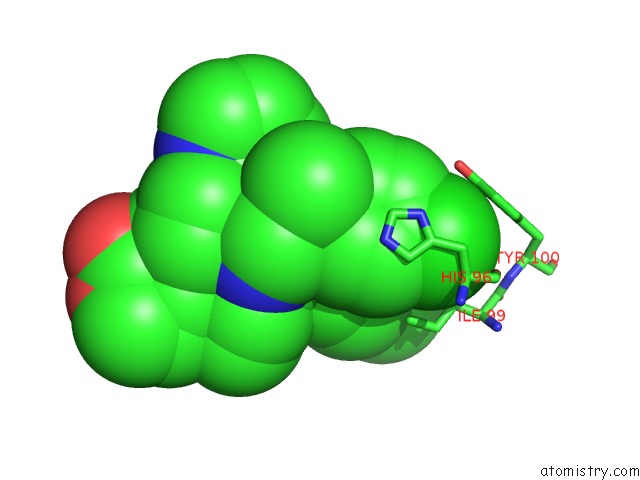
Mono view
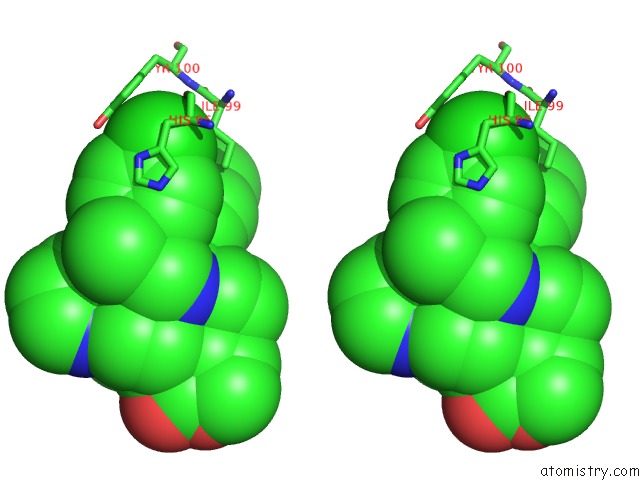
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of X-Ray Structure of the P53-MDM2 Inhibitor NMI801 Bound to HDM2 at 2.1A Resolution within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 6i29
Go back to
Chlorine binding site 2 out
of 2 in the X-Ray Structure of the P53-MDM2 Inhibitor NMI801 Bound to HDM2 at 2.1A Resolution
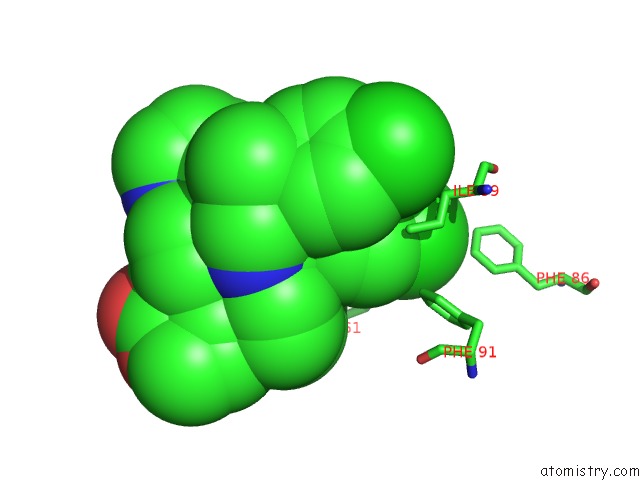
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of X-Ray Structure of the P53-MDM2 Inhibitor NMI801 Bound to HDM2 at 2.1A Resolution within 5.0Å range:
|
Reference:
J.Ornstein,
Y.Iwamoto,
M.Prytyskach,
M.Miller,
J.Kallen,
S.Ferretti,
P.Holzer,
W.Forrester,
R.Weissleder,
G.Lahav.
P53 Dynamics Vary Between Tissues and Are Linked with Radiation Sensitivity To Be Published.
Page generated: Sun Jul 28 01:31:50 2024
Last articles
Cl in 2WZNCl in 2WZX
Cl in 2WZD
Cl in 2WZQ
Cl in 2WYW
Cl in 2WYT
Cl in 2WZC
Cl in 2WUV
Cl in 2WZB
Cl in 2WYJ