Chlorine »
PDB 6lw2-6m7z »
6lx5 »
Chlorine in PDB 6lx5: X-Ray Structure of Human Pparalpha Ligand Binding Domain-Ciprofibrate Co-Crystals Obtained By Delipidation and Co-Crystallization
Protein crystallography data
The structure of X-Ray Structure of Human Pparalpha Ligand Binding Domain-Ciprofibrate Co-Crystals Obtained By Delipidation and Co-Crystallization, PDB code: 6lx5
was solved by
S.Kamata,
K.Saito,
A.Honda,
R.Ishikawa,
T.Oyama,
I.Ishii,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 36.41 / 1.87 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 60.585, 101.350, 61.745, 90.00, 100.73, 90.00 |
| R / Rfree (%) | 19.9 / 22.1 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the X-Ray Structure of Human Pparalpha Ligand Binding Domain-Ciprofibrate Co-Crystals Obtained By Delipidation and Co-Crystallization
(pdb code 6lx5). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 8 binding sites of Chlorine where determined in the X-Ray Structure of Human Pparalpha Ligand Binding Domain-Ciprofibrate Co-Crystals Obtained By Delipidation and Co-Crystallization, PDB code: 6lx5:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
In total 8 binding sites of Chlorine where determined in the X-Ray Structure of Human Pparalpha Ligand Binding Domain-Ciprofibrate Co-Crystals Obtained By Delipidation and Co-Crystallization, PDB code: 6lx5:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
Chlorine binding site 1 out of 8 in 6lx5
Go back to
Chlorine binding site 1 out
of 8 in the X-Ray Structure of Human Pparalpha Ligand Binding Domain-Ciprofibrate Co-Crystals Obtained By Delipidation and Co-Crystallization
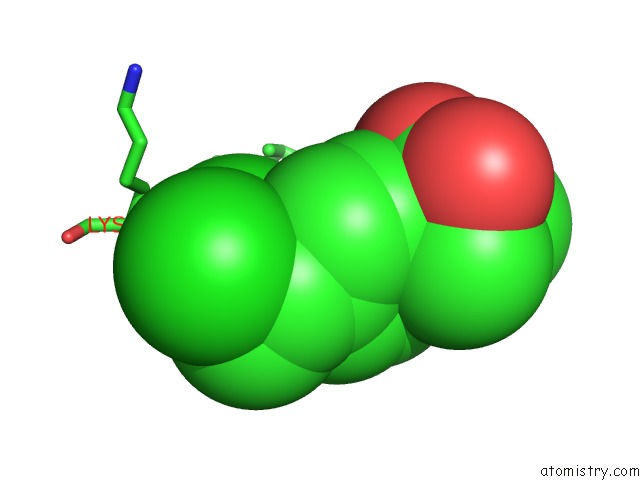
Mono view
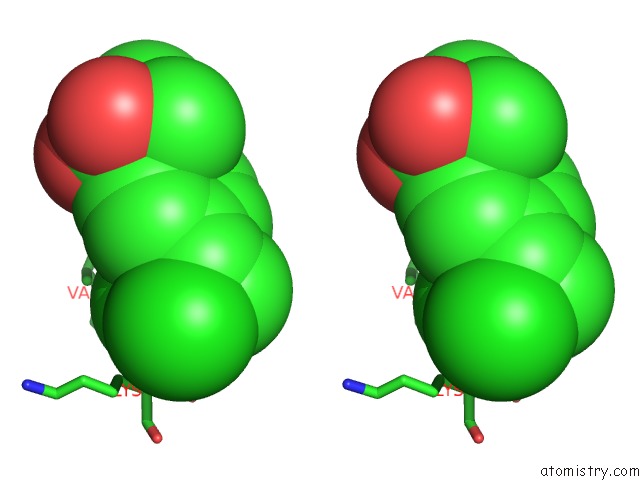
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of X-Ray Structure of Human Pparalpha Ligand Binding Domain-Ciprofibrate Co-Crystals Obtained By Delipidation and Co-Crystallization within 5.0Å range:
|
Chlorine binding site 2 out of 8 in 6lx5
Go back to
Chlorine binding site 2 out
of 8 in the X-Ray Structure of Human Pparalpha Ligand Binding Domain-Ciprofibrate Co-Crystals Obtained By Delipidation and Co-Crystallization
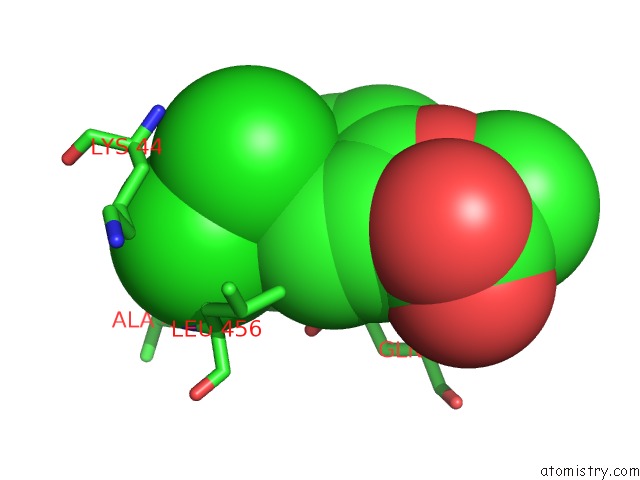
Mono view
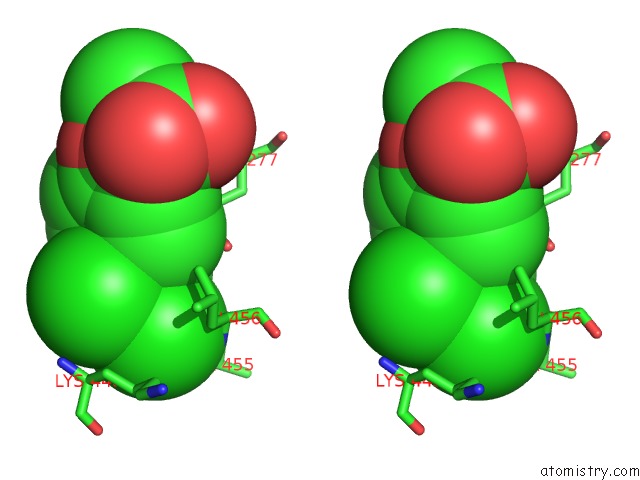
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of X-Ray Structure of Human Pparalpha Ligand Binding Domain-Ciprofibrate Co-Crystals Obtained By Delipidation and Co-Crystallization within 5.0Å range:
|
Chlorine binding site 3 out of 8 in 6lx5
Go back to
Chlorine binding site 3 out
of 8 in the X-Ray Structure of Human Pparalpha Ligand Binding Domain-Ciprofibrate Co-Crystals Obtained By Delipidation and Co-Crystallization
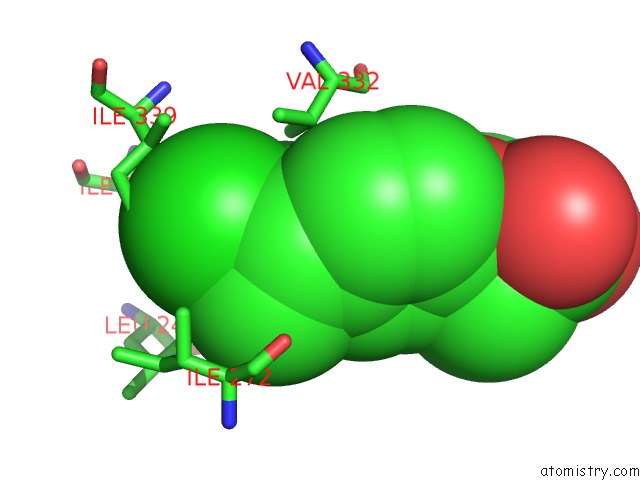
Mono view
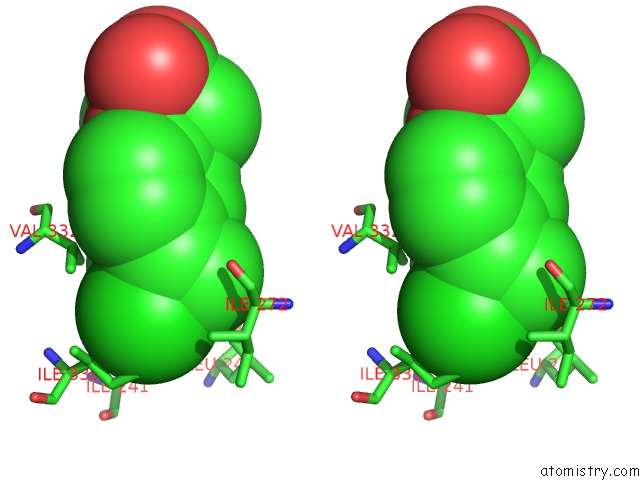
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of X-Ray Structure of Human Pparalpha Ligand Binding Domain-Ciprofibrate Co-Crystals Obtained By Delipidation and Co-Crystallization within 5.0Å range:
|
Chlorine binding site 4 out of 8 in 6lx5
Go back to
Chlorine binding site 4 out
of 8 in the X-Ray Structure of Human Pparalpha Ligand Binding Domain-Ciprofibrate Co-Crystals Obtained By Delipidation and Co-Crystallization
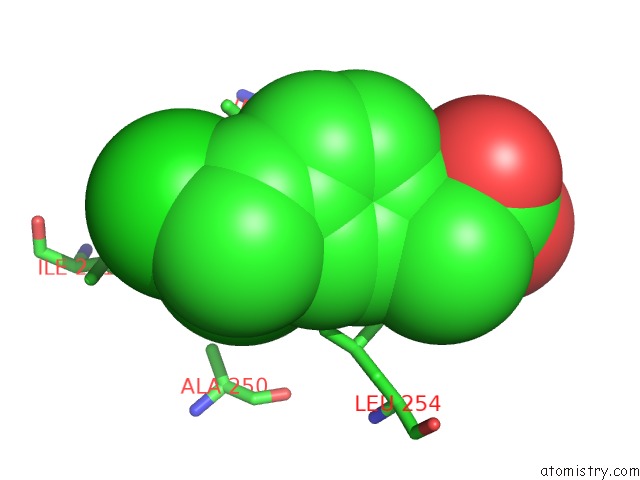
Mono view
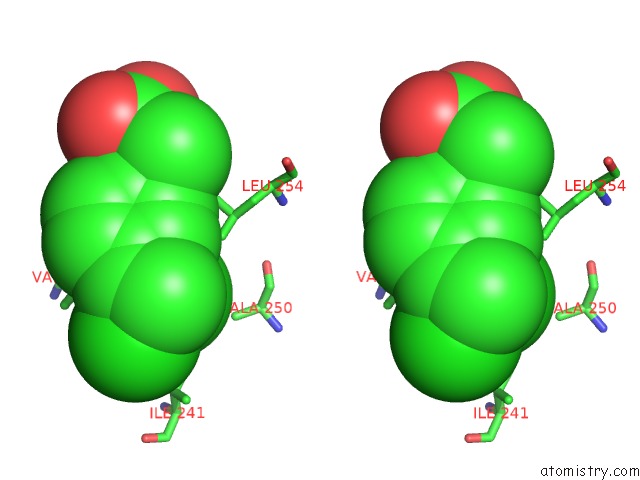
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of X-Ray Structure of Human Pparalpha Ligand Binding Domain-Ciprofibrate Co-Crystals Obtained By Delipidation and Co-Crystallization within 5.0Å range:
|
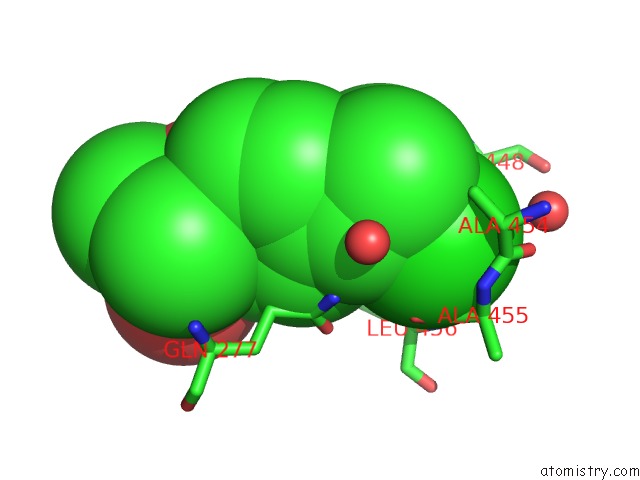
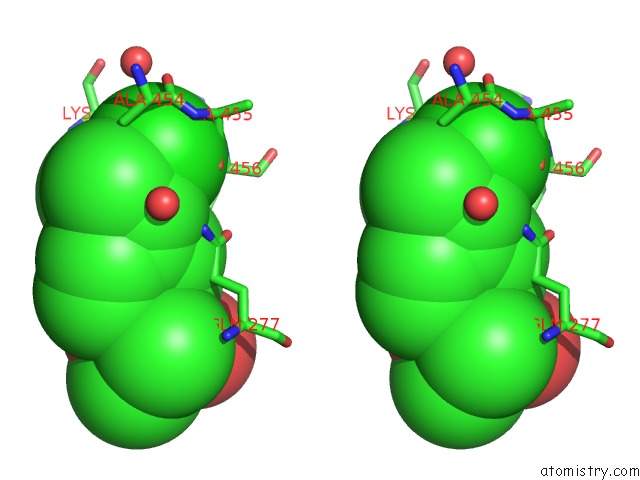
Chlorine binding site 5 out of 8 in 6lx5
Go back to
Chlorine binding site 5 out
of 8 in the X-Ray Structure of Human Pparalpha Ligand Binding Domain-Ciprofibrate Co-Crystals Obtained By Delipidation and Co-Crystallization
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of X-Ray Structure of Human Pparalpha Ligand Binding Domain-Ciprofibrate Co-Crystals Obtained By Delipidation and Co-Crystallization within 5.0Å range:
|
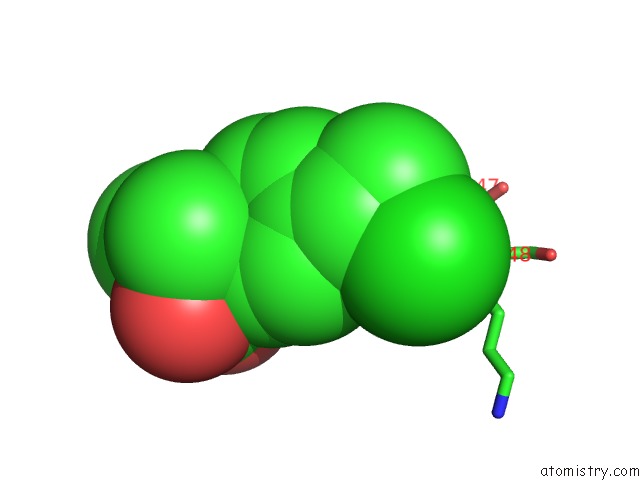
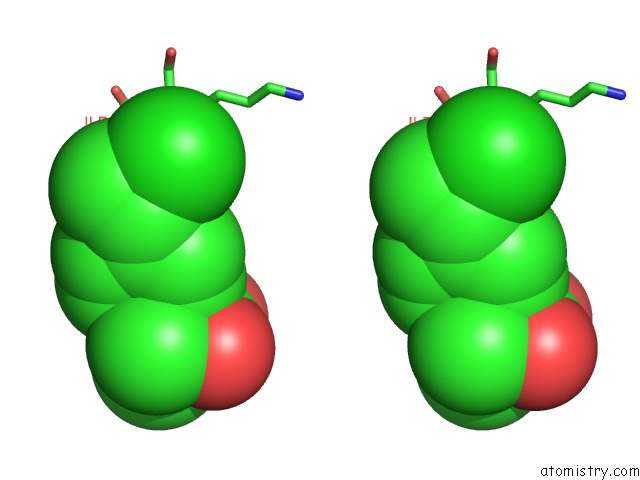
Chlorine binding site 6 out of 8 in 6lx5
Go back to
Chlorine binding site 6 out
of 8 in the X-Ray Structure of Human Pparalpha Ligand Binding Domain-Ciprofibrate Co-Crystals Obtained By Delipidation and Co-Crystallization
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of X-Ray Structure of Human Pparalpha Ligand Binding Domain-Ciprofibrate Co-Crystals Obtained By Delipidation and Co-Crystallization within 5.0Å range:
|
Chlorine binding site 7 out of 8 in 6lx5
Go back to
Chlorine binding site 7 out
of 8 in the X-Ray Structure of Human Pparalpha Ligand Binding Domain-Ciprofibrate Co-Crystals Obtained By Delipidation and Co-Crystallization
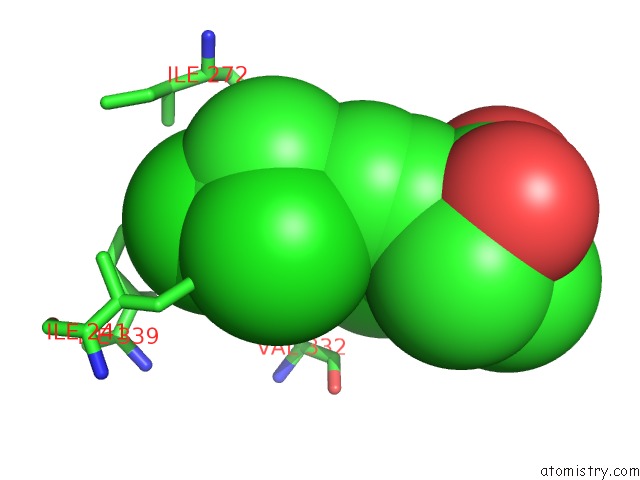
Mono view
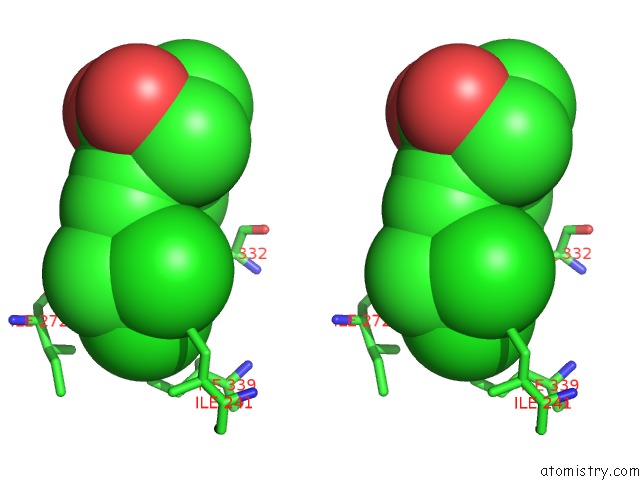
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 7 of X-Ray Structure of Human Pparalpha Ligand Binding Domain-Ciprofibrate Co-Crystals Obtained By Delipidation and Co-Crystallization within 5.0Å range:
|
Chlorine binding site 8 out of 8 in 6lx5
Go back to
Chlorine binding site 8 out
of 8 in the X-Ray Structure of Human Pparalpha Ligand Binding Domain-Ciprofibrate Co-Crystals Obtained By Delipidation and Co-Crystallization
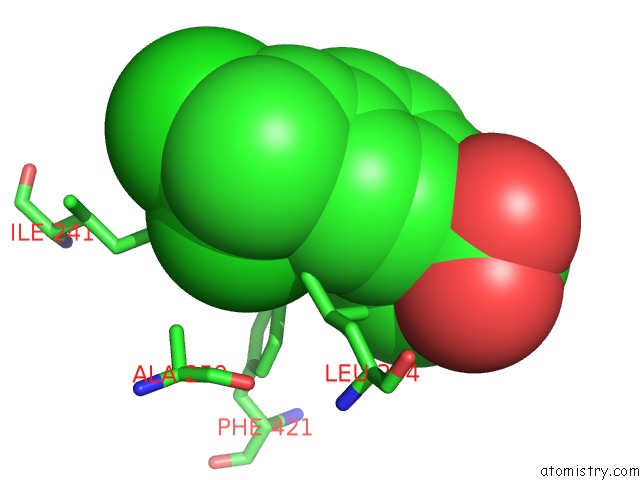
Mono view
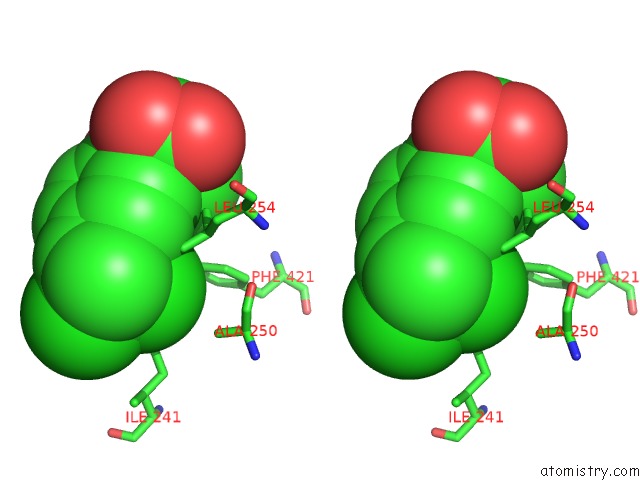
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 8 of X-Ray Structure of Human Pparalpha Ligand Binding Domain-Ciprofibrate Co-Crystals Obtained By Delipidation and Co-Crystallization within 5.0Å range:
|
Reference:
S.Kamata,
T.Oyama,
K.Saito,
A.Honda,
Y.Yamamoto,
K.Suda,
R.Ishikawa,
T.Itoh,
Y.Watanabe,
T.Shibata,
K.Uchida,
M.Suematsu,
I.Ishii.
Ppar Alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates. Iscience V. 23 01727 2020.
ISSN: ESSN 2589-0042
PubMed: 33205029
DOI: 10.1016/J.ISCI.2020.101727
Page generated: Sat Jul 12 16:37:04 2025
ISSN: ESSN 2589-0042
PubMed: 33205029
DOI: 10.1016/J.ISCI.2020.101727
Last articles
Cl in 6SLRCl in 6SLO
Cl in 6SLV
Cl in 6SKH
Cl in 6SIS
Cl in 6SJZ
Cl in 6SK8
Cl in 6SJ7
Cl in 6SIQ
Cl in 6SIM