Chlorine »
PDB 6lw2-6m7z »
6m12 »
Chlorine in PDB 6m12: Crystal Structure of Rnase L in Complex with SU11652
Protein crystallography data
The structure of Crystal Structure of Rnase L in Complex with SU11652, PDB code: 6m12
was solved by
J.Tang,
H.Huang,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 57.30 / 2.40 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 58.710, 110.380, 262.590, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 23.1 / 26.6 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of Rnase L in Complex with SU11652
(pdb code 6m12). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Crystal Structure of Rnase L in Complex with SU11652, PDB code: 6m12:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Crystal Structure of Rnase L in Complex with SU11652, PDB code: 6m12:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 6m12
Go back to
Chlorine binding site 1 out
of 2 in the Crystal Structure of Rnase L in Complex with SU11652
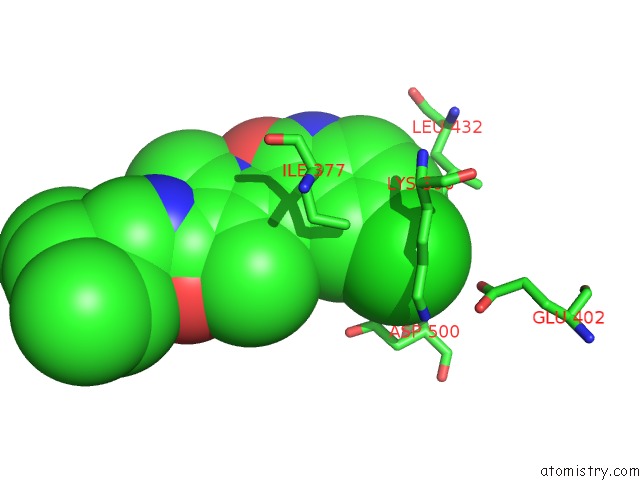
Mono view
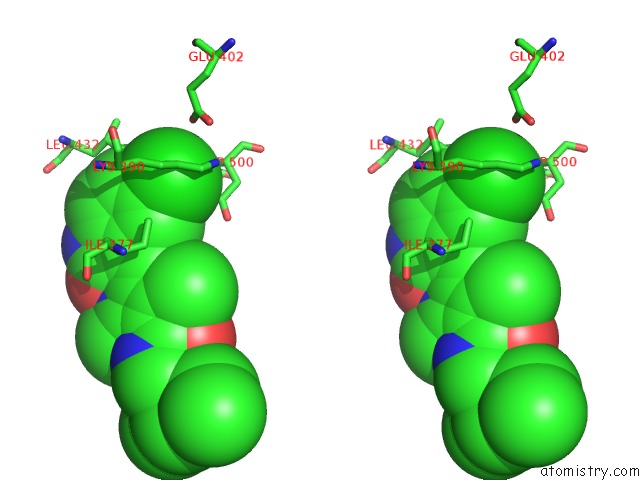
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Rnase L in Complex with SU11652 within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 6m12
Go back to
Chlorine binding site 2 out
of 2 in the Crystal Structure of Rnase L in Complex with SU11652
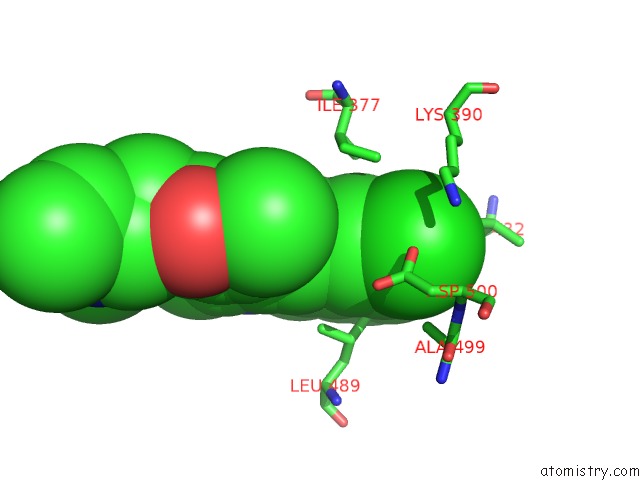
Mono view
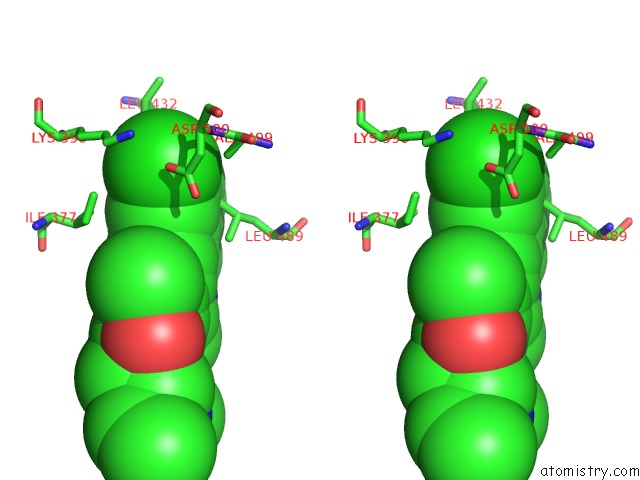
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of Rnase L in Complex with SU11652 within 5.0Å range:
|
Reference:
J.Tang,
Y.Wang,
H.Zhou,
Y.Ye,
M.Talukdar,
Z.Fu,
Z.Liu,
J.Li,
D.Neculai,
J.Gao,
H.Huang.
Sunitinib Inhibits Rnase L By Destabilizing Its Active Dimer Conformation. Biochem.J. V. 477 3387 2020.
ISSN: ESSN 1470-8728
PubMed: 32830849
DOI: 10.1042/BCJ20200260
Page generated: Sat Jul 12 16:38:55 2025
ISSN: ESSN 1470-8728
PubMed: 32830849
DOI: 10.1042/BCJ20200260
Last articles
Cl in 6XCXCl in 6XCF
Cl in 6XC0
Cl in 6XCE
Cl in 6X9N
Cl in 6XBV
Cl in 6XBS
Cl in 6XBR
Cl in 6XBL
Cl in 6X9F