Chlorine »
PDB 6p2l-6p8z »
6p2o »
Chlorine in PDB 6p2o: Crystal Structure of Streptomyces Rapamycinicus GH74 in Complex with Xyloglucan Fragments Xllg and Xxxg
Protein crystallography data
The structure of Crystal Structure of Streptomyces Rapamycinicus GH74 in Complex with Xyloglucan Fragments Xllg and Xxxg, PDB code: 6p2o
was solved by
P.J.Stogios,
T.Skarina,
G.Arnal,
H.Brumer,
A.Savchenko,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 24.82 / 1.88 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 92.996, 112.788, 222.759, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 14.8 / 18 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of Streptomyces Rapamycinicus GH74 in Complex with Xyloglucan Fragments Xllg and Xxxg
(pdb code 6p2o). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 7 binding sites of Chlorine where determined in the Crystal Structure of Streptomyces Rapamycinicus GH74 in Complex with Xyloglucan Fragments Xllg and Xxxg, PDB code: 6p2o:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7;
In total 7 binding sites of Chlorine where determined in the Crystal Structure of Streptomyces Rapamycinicus GH74 in Complex with Xyloglucan Fragments Xllg and Xxxg, PDB code: 6p2o:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7;
Chlorine binding site 1 out of 7 in 6p2o
Go back to
Chlorine binding site 1 out
of 7 in the Crystal Structure of Streptomyces Rapamycinicus GH74 in Complex with Xyloglucan Fragments Xllg and Xxxg
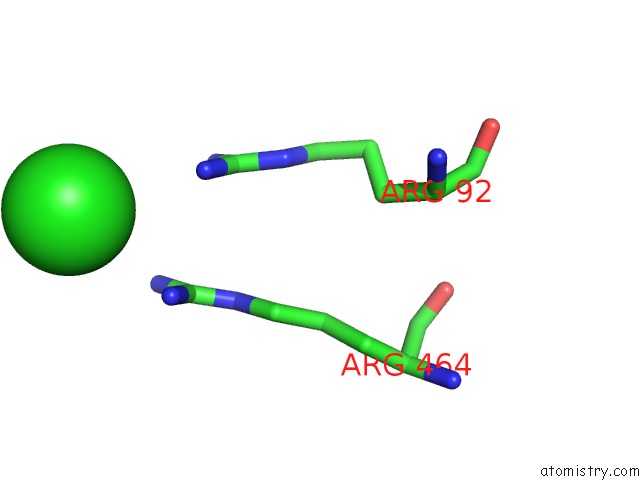
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Streptomyces Rapamycinicus GH74 in Complex with Xyloglucan Fragments Xllg and Xxxg within 5.0Å range:
|
Chlorine binding site 2 out of 7 in 6p2o
Go back to
Chlorine binding site 2 out
of 7 in the Crystal Structure of Streptomyces Rapamycinicus GH74 in Complex with Xyloglucan Fragments Xllg and Xxxg
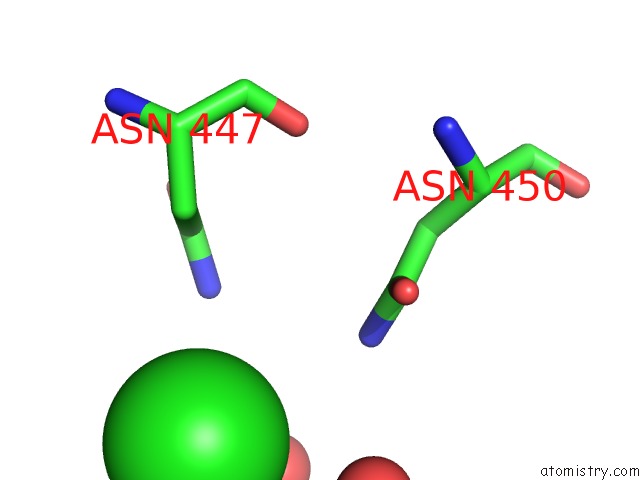
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of Streptomyces Rapamycinicus GH74 in Complex with Xyloglucan Fragments Xllg and Xxxg within 5.0Å range:
|
Chlorine binding site 3 out of 7 in 6p2o
Go back to
Chlorine binding site 3 out
of 7 in the Crystal Structure of Streptomyces Rapamycinicus GH74 in Complex with Xyloglucan Fragments Xllg and Xxxg

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Crystal Structure of Streptomyces Rapamycinicus GH74 in Complex with Xyloglucan Fragments Xllg and Xxxg within 5.0Å range:
|
Chlorine binding site 4 out of 7 in 6p2o
Go back to
Chlorine binding site 4 out
of 7 in the Crystal Structure of Streptomyces Rapamycinicus GH74 in Complex with Xyloglucan Fragments Xllg and Xxxg

Mono view
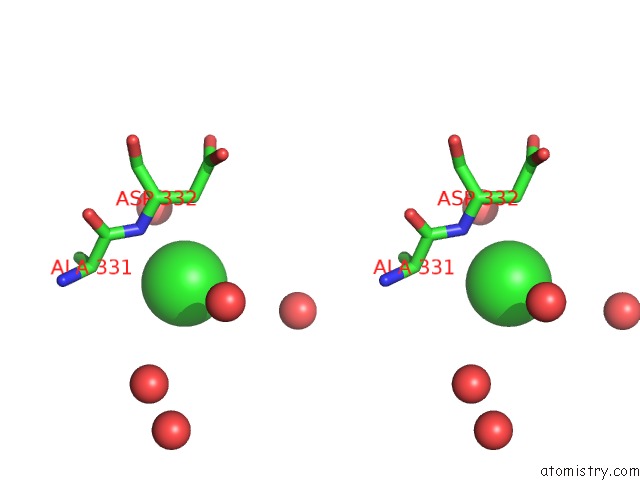
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Crystal Structure of Streptomyces Rapamycinicus GH74 in Complex with Xyloglucan Fragments Xllg and Xxxg within 5.0Å range:
|
Chlorine binding site 5 out of 7 in 6p2o
Go back to
Chlorine binding site 5 out
of 7 in the Crystal Structure of Streptomyces Rapamycinicus GH74 in Complex with Xyloglucan Fragments Xllg and Xxxg
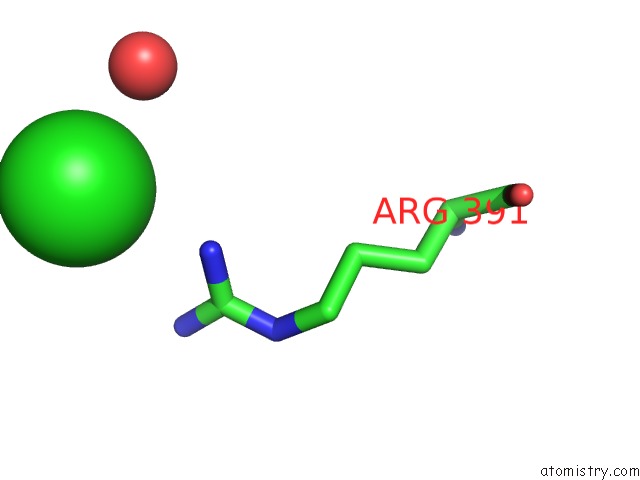
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Crystal Structure of Streptomyces Rapamycinicus GH74 in Complex with Xyloglucan Fragments Xllg and Xxxg within 5.0Å range:
|
Chlorine binding site 6 out of 7 in 6p2o
Go back to
Chlorine binding site 6 out
of 7 in the Crystal Structure of Streptomyces Rapamycinicus GH74 in Complex with Xyloglucan Fragments Xllg and Xxxg

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of Crystal Structure of Streptomyces Rapamycinicus GH74 in Complex with Xyloglucan Fragments Xllg and Xxxg within 5.0Å range:
|
Chlorine binding site 7 out of 7 in 6p2o
Go back to
Chlorine binding site 7 out
of 7 in the Crystal Structure of Streptomyces Rapamycinicus GH74 in Complex with Xyloglucan Fragments Xllg and Xxxg

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 7 of Crystal Structure of Streptomyces Rapamycinicus GH74 in Complex with Xyloglucan Fragments Xllg and Xxxg within 5.0Å range:
|
Reference:
G.Arnal,
P.J.Stogios,
J.Asohan,
M.A.Attia,
T.Skarina,
A.H.Viborg,
B.Henrissat,
A.Savchenko,
H.Brumer.
Substrate Specificity, Regiospecificity, and Processivity in Glycoside Hydrolase Family 74. J.Biol.Chem. V. 294 13233 2019.
ISSN: ESSN 1083-351X
PubMed: 31324716
DOI: 10.1074/JBC.RA119.009861
Page generated: Sat Jul 12 18:09:48 2025
ISSN: ESSN 1083-351X
PubMed: 31324716
DOI: 10.1074/JBC.RA119.009861
Last articles
Cl in 8B2KCl in 8B10
Cl in 8B2D
Cl in 8B2I
Cl in 8B08
Cl in 8B05
Cl in 8B06
Cl in 8B03
Cl in 8AYQ
Cl in 8B02