Chlorine »
PDB 6tdq-6tma »
6tdq »
Chlorine in PDB 6tdq: Crystal Structure of the Disulfide Engineered Hla-A0201 Molecule in Complex with One Gm Dipeptide in the A Pocket and One Gm Dipeptide in the F Pocket.
Protein crystallography data
The structure of Crystal Structure of the Disulfide Engineered Hla-A0201 Molecule in Complex with One Gm Dipeptide in the A Pocket and One Gm Dipeptide in the F Pocket., PDB code: 6tdq
was solved by
R.Anjanappa,
M.Garcia Alai,
S.Springer,
R.Meijers,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 83.72 / 1.60 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 58.643, 84.124, 83.716, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 16.9 / 21.4 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of the Disulfide Engineered Hla-A0201 Molecule in Complex with One Gm Dipeptide in the A Pocket and One Gm Dipeptide in the F Pocket.
(pdb code 6tdq). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Crystal Structure of the Disulfide Engineered Hla-A0201 Molecule in Complex with One Gm Dipeptide in the A Pocket and One Gm Dipeptide in the F Pocket., PDB code: 6tdq:
In total only one binding site of Chlorine was determined in the Crystal Structure of the Disulfide Engineered Hla-A0201 Molecule in Complex with One Gm Dipeptide in the A Pocket and One Gm Dipeptide in the F Pocket., PDB code: 6tdq:
Chlorine binding site 1 out of 1 in 6tdq
Go back to
Chlorine binding site 1 out
of 1 in the Crystal Structure of the Disulfide Engineered Hla-A0201 Molecule in Complex with One Gm Dipeptide in the A Pocket and One Gm Dipeptide in the F Pocket.
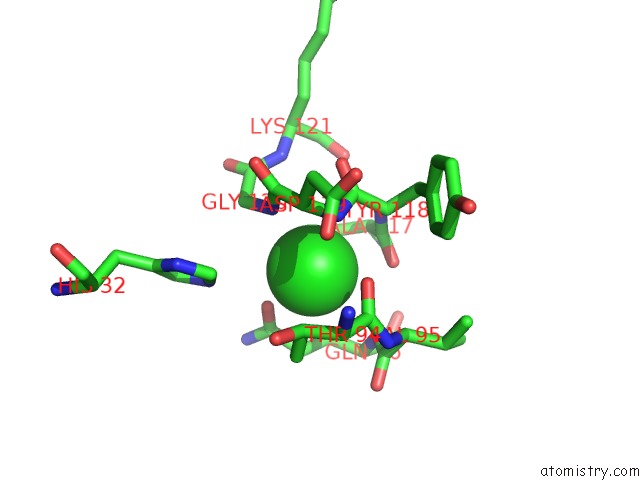
Mono view
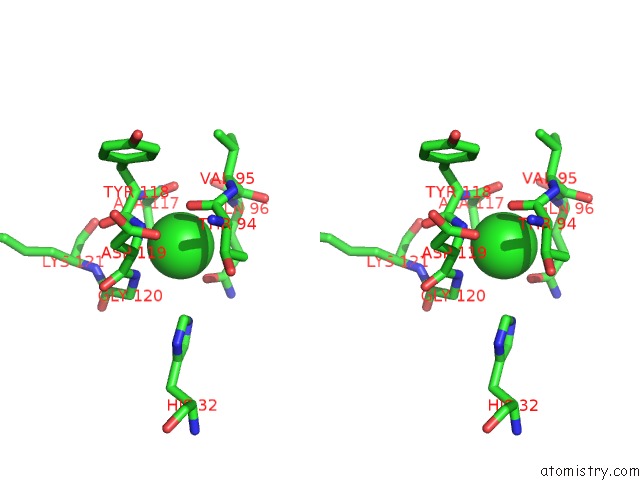
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of the Disulfide Engineered Hla-A0201 Molecule in Complex with One Gm Dipeptide in the A Pocket and One Gm Dipeptide in the F Pocket. within 5.0Å range:
|
Reference:
R.Anjanappa,
M.Garcia-Alai,
J.D.Kopicki,
J.Lockhauserbaumer,
M.Aboelmagd,
J.Hinrichs,
I.M.Nemtanu,
C.Uetrecht,
M.Zacharias,
S.Springer,
R.Meijers.
Structures of Peptide-Free and Partially Loaded Mhc Class I Molecules Reveal Mechanisms of Peptide Selection. Nat Commun V. 11 1314 2020.
ISSN: ESSN 2041-1723
PubMed: 32161266
DOI: 10.1038/S41467-020-14862-4
Page generated: Sat Jul 12 20:06:51 2025
ISSN: ESSN 2041-1723
PubMed: 32161266
DOI: 10.1038/S41467-020-14862-4
Last articles
Cl in 8BYHCl in 8BZ4
Cl in 8BYY
Cl in 8BYG
Cl in 8BYO
Cl in 8BYK
Cl in 8BYC
Cl in 8BYF
Cl in 8BYE
Cl in 8BYD