Chlorine »
PDB 2kug-2nwh »
2mgy »
Chlorine in PDB 2mgy: Solution Structure of the Mitochondrial Translocator Protein (Tspo) in Complex with Its High-Affinity Ligand PK11195
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Solution Structure of the Mitochondrial Translocator Protein (Tspo) in Complex with Its High-Affinity Ligand PK11195
(pdb code 2mgy). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Solution Structure of the Mitochondrial Translocator Protein (Tspo) in Complex with Its High-Affinity Ligand PK11195, PDB code: 2mgy:
In total only one binding site of Chlorine was determined in the Solution Structure of the Mitochondrial Translocator Protein (Tspo) in Complex with Its High-Affinity Ligand PK11195, PDB code: 2mgy:
Chlorine binding site 1 out of 1 in 2mgy
Go back to
Chlorine binding site 1 out
of 1 in the Solution Structure of the Mitochondrial Translocator Protein (Tspo) in Complex with Its High-Affinity Ligand PK11195
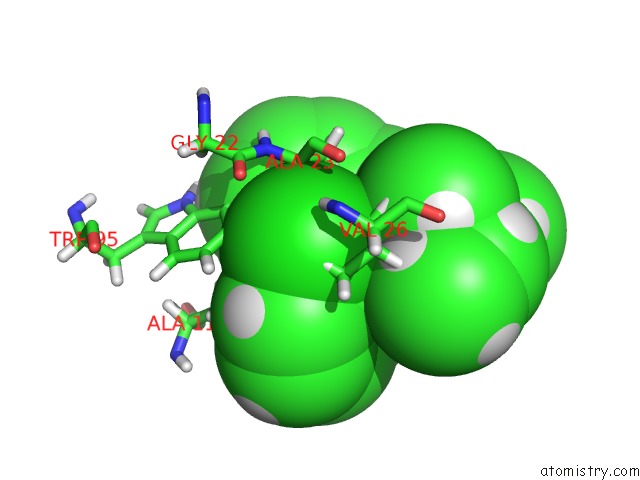
Mono view
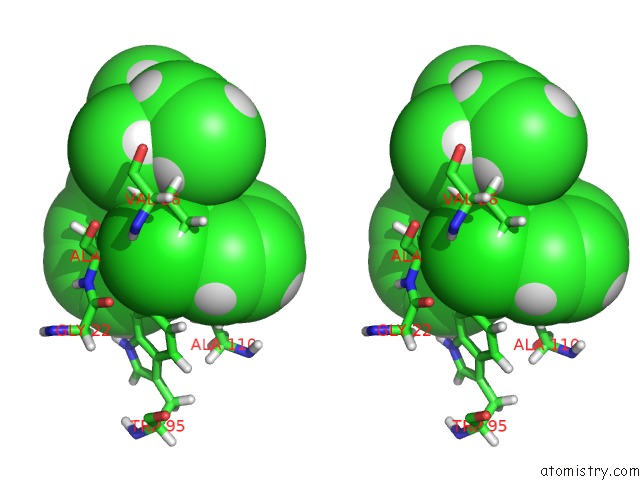
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Solution Structure of the Mitochondrial Translocator Protein (Tspo) in Complex with Its High-Affinity Ligand PK11195 within 5.0Å range:
|
Reference:
L.Jaremko,
M.Jaremko,
K.Giller,
S.Becker,
M.Zweckstetter.
Structure of the Mitochondrial Translocator Protein in Complex with A Diagnostic Ligand. Science V. 343 1363 2014.
ISSN: ISSN 0036-8075
PubMed: 24653034
DOI: 10.1126/SCIENCE.1248725
Page generated: Thu Jul 10 23:17:26 2025
ISSN: ISSN 0036-8075
PubMed: 24653034
DOI: 10.1126/SCIENCE.1248725
Last articles
Cl in 3D15Cl in 3D0S
Cl in 3D0I
Cl in 3D0H
Cl in 3D04
Cl in 3D0G
Cl in 3CZ2
Cl in 3D02
Cl in 3CZ1
Cl in 3D00