Chlorine »
PDB 2kug-2nwh »
2n02 »
Chlorine in PDB 2n02: Solution Structure of the A147T Variant of the Mitochondrial Translocator Protein (Tspo) in Complex with PK11195
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Solution Structure of the A147T Variant of the Mitochondrial Translocator Protein (Tspo) in Complex with PK11195
(pdb code 2n02). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Solution Structure of the A147T Variant of the Mitochondrial Translocator Protein (Tspo) in Complex with PK11195, PDB code: 2n02:
In total only one binding site of Chlorine was determined in the Solution Structure of the A147T Variant of the Mitochondrial Translocator Protein (Tspo) in Complex with PK11195, PDB code: 2n02:
Chlorine binding site 1 out of 1 in 2n02
Go back to
Chlorine binding site 1 out
of 1 in the Solution Structure of the A147T Variant of the Mitochondrial Translocator Protein (Tspo) in Complex with PK11195
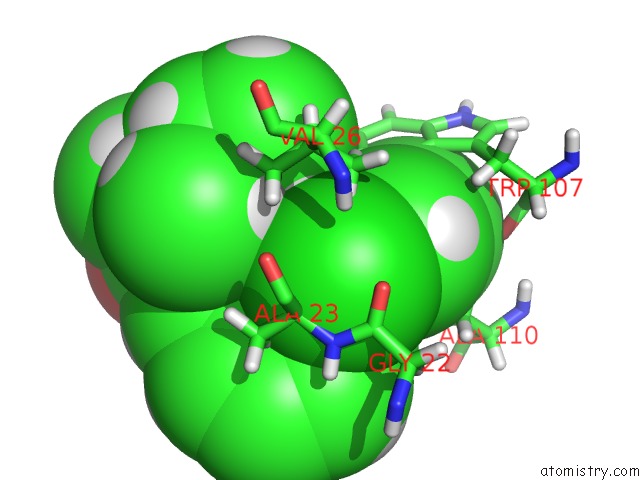
Mono view
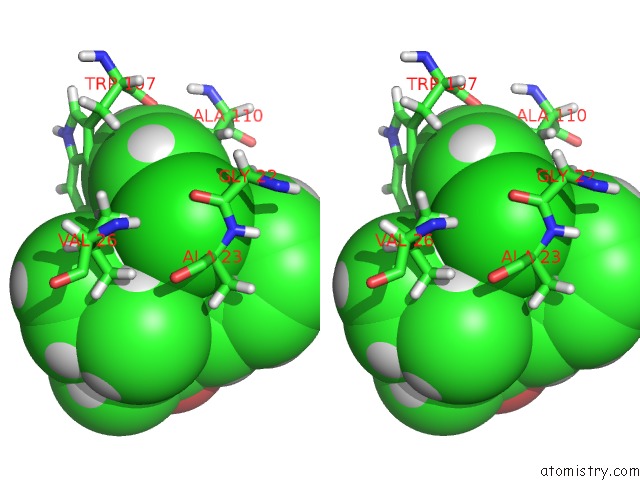
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Solution Structure of the A147T Variant of the Mitochondrial Translocator Protein (Tspo) in Complex with PK11195 within 5.0Å range:
|
Reference:
M.Jaremko,
M.Jaremko,
K.Giller,
S.Becker,
M.Zweckstetter.
Structural Integrity of the A147T Polymorph of Mammalian Tspo. Chembiochem V. 16 1483 2015.
ISSN: ISSN 1439-4227
PubMed: 25974690
DOI: 10.1002/CBIC.201500217
Page generated: Thu Jul 10 23:17:37 2025
ISSN: ISSN 1439-4227
PubMed: 25974690
DOI: 10.1002/CBIC.201500217
Last articles
Cl in 3CC4Cl in 3CC7
Cl in 3CC2
Cl in 3CCC
Cl in 3C8V
Cl in 3C7T
Cl in 3CBX
Cl in 3C9I
Cl in 3CAJ
Cl in 3C84