Chlorine »
PDB 2xev-2xmc »
2xfg »
Chlorine in PDB 2xfg: Reassembly and Co-Crystallization of A Family 9 Processive Endoglucanase From Separately Expressed GH9 and CBM3C Modules
Enzymatic activity of Reassembly and Co-Crystallization of A Family 9 Processive Endoglucanase From Separately Expressed GH9 and CBM3C Modules
All present enzymatic activity of Reassembly and Co-Crystallization of A Family 9 Processive Endoglucanase From Separately Expressed GH9 and CBM3C Modules:
3.2.1.4;
3.2.1.4;
Protein crystallography data
The structure of Reassembly and Co-Crystallization of A Family 9 Processive Endoglucanase From Separately Expressed GH9 and CBM3C Modules, PDB code: 2xfg
was solved by
S.Petkun,
R.Lamed,
S.Jindou,
T.Burstein,
O.Yaniv,
Y.Shoham,
J.W.L.Shimon,
E.A.Bayer,
F.Frolow,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20.868 / 1.68 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 70.399, 88.541, 106.486, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 14.14 / 17.59 |
Other elements in 2xfg:
The structure of Reassembly and Co-Crystallization of A Family 9 Processive Endoglucanase From Separately Expressed GH9 and CBM3C Modules also contains other interesting chemical elements:
| Calcium | (Ca) | 2 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Reassembly and Co-Crystallization of A Family 9 Processive Endoglucanase From Separately Expressed GH9 and CBM3C Modules
(pdb code 2xfg). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 3 binding sites of Chlorine where determined in the Reassembly and Co-Crystallization of A Family 9 Processive Endoglucanase From Separately Expressed GH9 and CBM3C Modules, PDB code: 2xfg:
Jump to Chlorine binding site number: 1; 2; 3;
In total 3 binding sites of Chlorine where determined in the Reassembly and Co-Crystallization of A Family 9 Processive Endoglucanase From Separately Expressed GH9 and CBM3C Modules, PDB code: 2xfg:
Jump to Chlorine binding site number: 1; 2; 3;
Chlorine binding site 1 out of 3 in 2xfg
Go back to
Chlorine binding site 1 out
of 3 in the Reassembly and Co-Crystallization of A Family 9 Processive Endoglucanase From Separately Expressed GH9 and CBM3C Modules
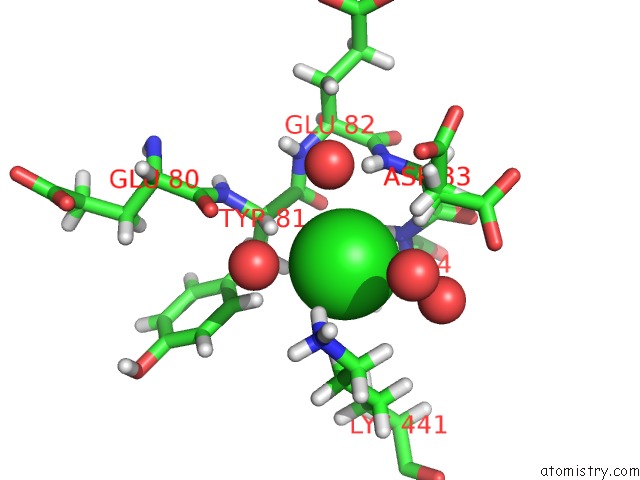
Mono view
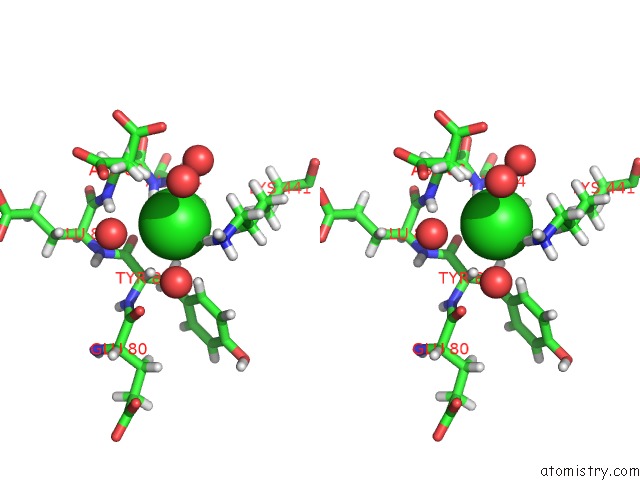
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Reassembly and Co-Crystallization of A Family 9 Processive Endoglucanase From Separately Expressed GH9 and CBM3C Modules within 5.0Å range:
|
Chlorine binding site 2 out of 3 in 2xfg
Go back to
Chlorine binding site 2 out
of 3 in the Reassembly and Co-Crystallization of A Family 9 Processive Endoglucanase From Separately Expressed GH9 and CBM3C Modules
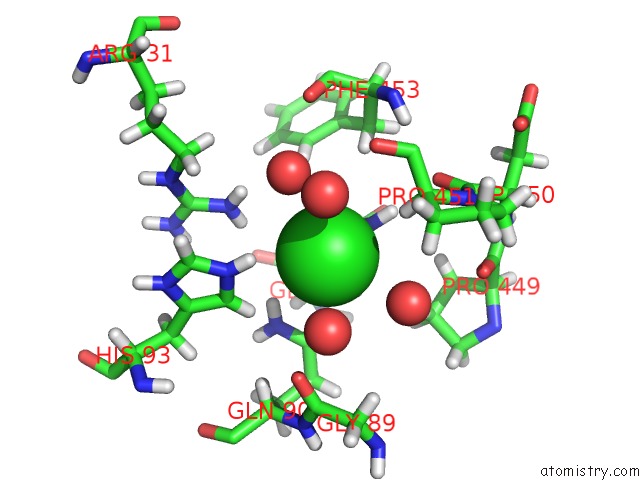
Mono view
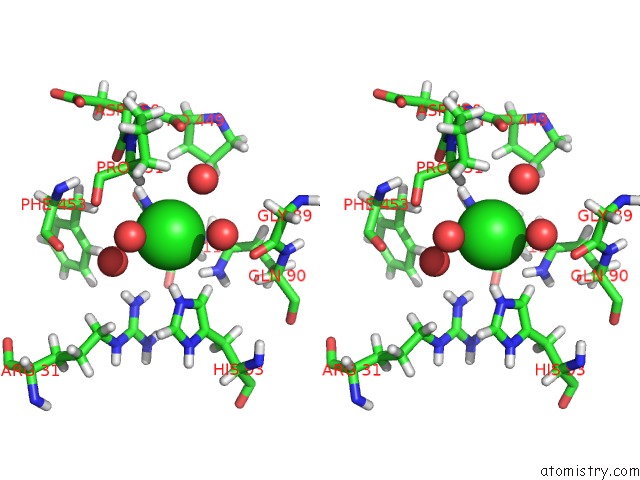
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Reassembly and Co-Crystallization of A Family 9 Processive Endoglucanase From Separately Expressed GH9 and CBM3C Modules within 5.0Å range:
|
Chlorine binding site 3 out of 3 in 2xfg
Go back to
Chlorine binding site 3 out
of 3 in the Reassembly and Co-Crystallization of A Family 9 Processive Endoglucanase From Separately Expressed GH9 and CBM3C Modules
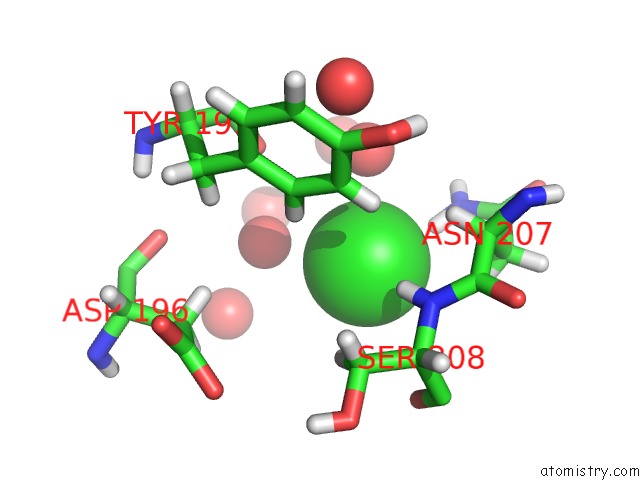
Mono view
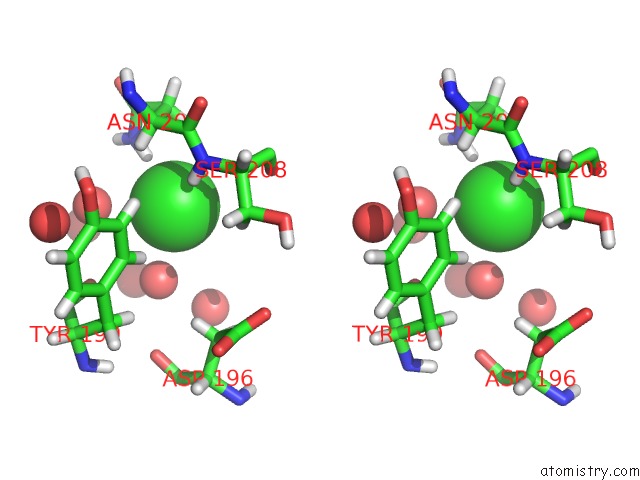
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Reassembly and Co-Crystallization of A Family 9 Processive Endoglucanase From Separately Expressed GH9 and CBM3C Modules within 5.0Å range:
|
Reference:
S.Petkun,
I.R.Grinberg,
R.Lamed,
S.Jindou,
T.Burstein,
O.Yaniv,
Y.Shoham,
J.W.L.Shimon,
E.A.Bayer,
F.Frolow.
Reassembly and Co-Crystallization of A Family 9 Processive Endoglucanase From Its Component Parts: Structural and Functional Significance of Intermodular Linker Peerj V. 3 E1126 2015.
ISSN: ISSN 2167-8359
PubMed: 26401442
DOI: 10.7717/PEERJ.1126
Page generated: Fri Jul 11 01:56:26 2025
ISSN: ISSN 2167-8359
PubMed: 26401442
DOI: 10.7717/PEERJ.1126
Last articles
Cl in 5UQPCl in 5UQF
Cl in 5UPZ
Cl in 5UPR
Cl in 5UQA
Cl in 5UPQ
Cl in 5UPT
Cl in 5UPY
Cl in 5UPG
Cl in 5UP7