Chlorine »
PDB 3pnf-3q12 »
3ppc »
Chlorine in PDB 3ppc: Crystal Structure of the Candida Albicans Methionine Synthase By Surface Entropy Reduction, Tyrosine Variant with Zinc
Enzymatic activity of Crystal Structure of the Candida Albicans Methionine Synthase By Surface Entropy Reduction, Tyrosine Variant with Zinc
All present enzymatic activity of Crystal Structure of the Candida Albicans Methionine Synthase By Surface Entropy Reduction, Tyrosine Variant with Zinc:
2.1.1.14;
2.1.1.14;
Protein crystallography data
The structure of Crystal Structure of the Candida Albicans Methionine Synthase By Surface Entropy Reduction, Tyrosine Variant with Zinc, PDB code: 3ppc
was solved by
D.Ubhi,
K.Kavanagh,
A.F.Monzingo,
J.D.Robertus,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 2.20 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 77.676, 92.685, 190.970, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 18.2 / 23.1 |
Other elements in 3ppc:
The structure of Crystal Structure of the Candida Albicans Methionine Synthase By Surface Entropy Reduction, Tyrosine Variant with Zinc also contains other interesting chemical elements:
| Zinc | (Zn) | 2 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of the Candida Albicans Methionine Synthase By Surface Entropy Reduction, Tyrosine Variant with Zinc
(pdb code 3ppc). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 8 binding sites of Chlorine where determined in the Crystal Structure of the Candida Albicans Methionine Synthase By Surface Entropy Reduction, Tyrosine Variant with Zinc, PDB code: 3ppc:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
In total 8 binding sites of Chlorine where determined in the Crystal Structure of the Candida Albicans Methionine Synthase By Surface Entropy Reduction, Tyrosine Variant with Zinc, PDB code: 3ppc:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
Chlorine binding site 1 out of 8 in 3ppc
Go back to
Chlorine binding site 1 out
of 8 in the Crystal Structure of the Candida Albicans Methionine Synthase By Surface Entropy Reduction, Tyrosine Variant with Zinc
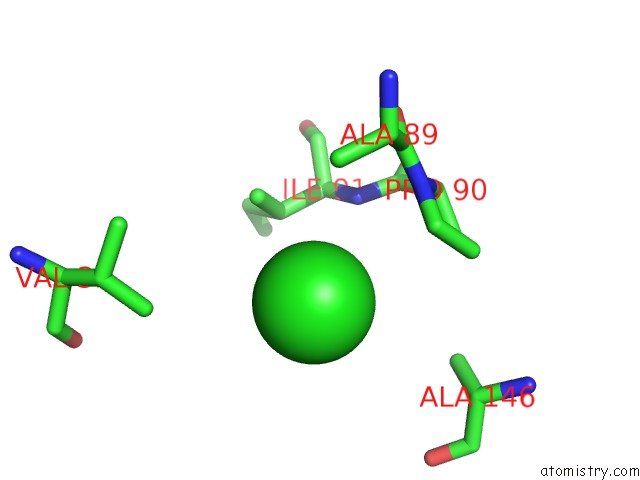
Mono view
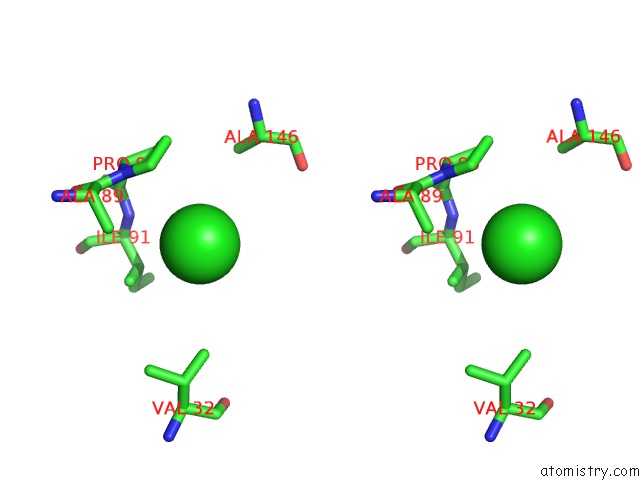
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of the Candida Albicans Methionine Synthase By Surface Entropy Reduction, Tyrosine Variant with Zinc within 5.0Å range:
|
Chlorine binding site 2 out of 8 in 3ppc
Go back to
Chlorine binding site 2 out
of 8 in the Crystal Structure of the Candida Albicans Methionine Synthase By Surface Entropy Reduction, Tyrosine Variant with Zinc
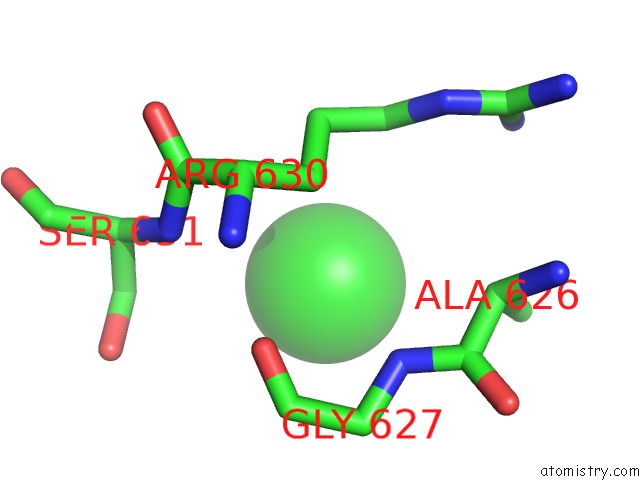
Mono view
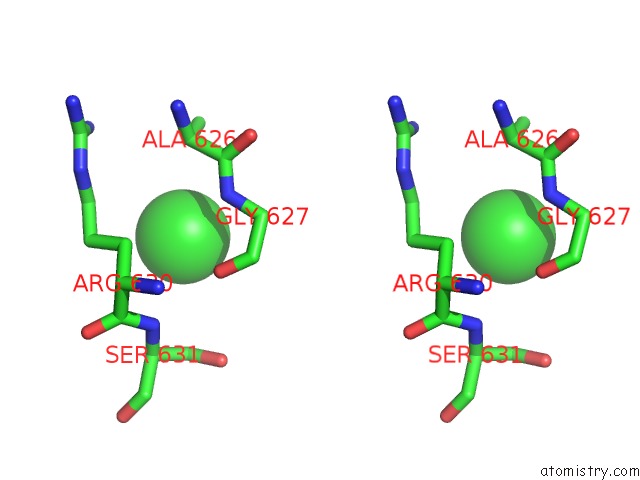
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of the Candida Albicans Methionine Synthase By Surface Entropy Reduction, Tyrosine Variant with Zinc within 5.0Å range:
|
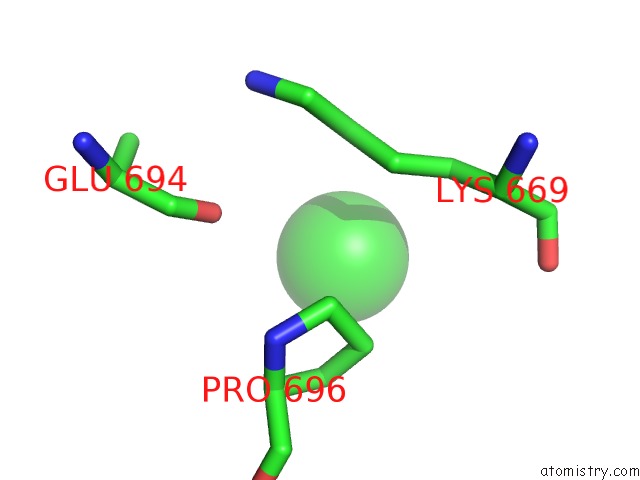
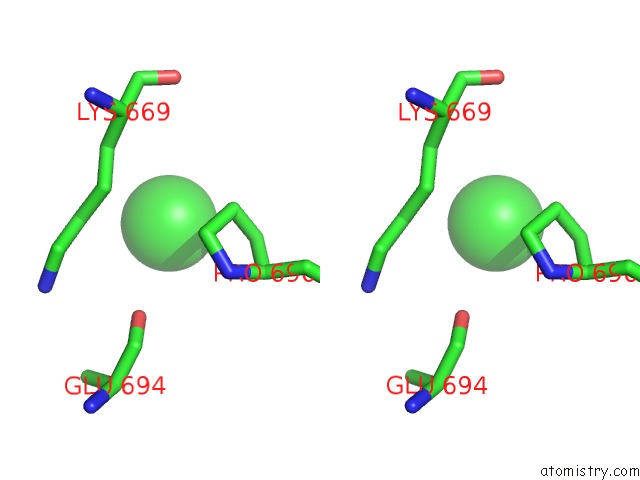
Chlorine binding site 3 out of 8 in 3ppc
Go back to
Chlorine binding site 3 out
of 8 in the Crystal Structure of the Candida Albicans Methionine Synthase By Surface Entropy Reduction, Tyrosine Variant with Zinc
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Crystal Structure of the Candida Albicans Methionine Synthase By Surface Entropy Reduction, Tyrosine Variant with Zinc within 5.0Å range:
|
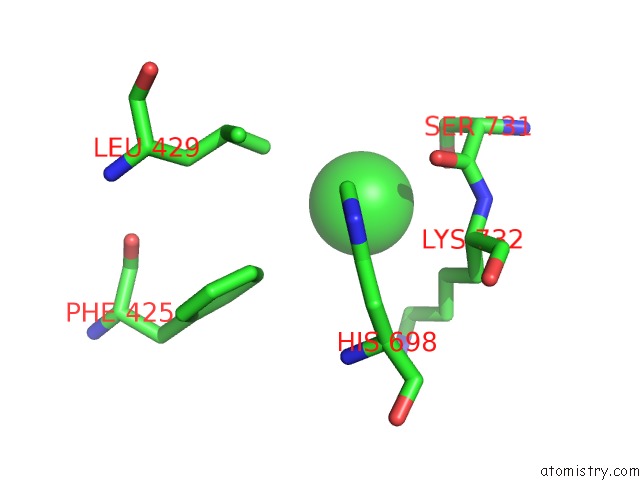
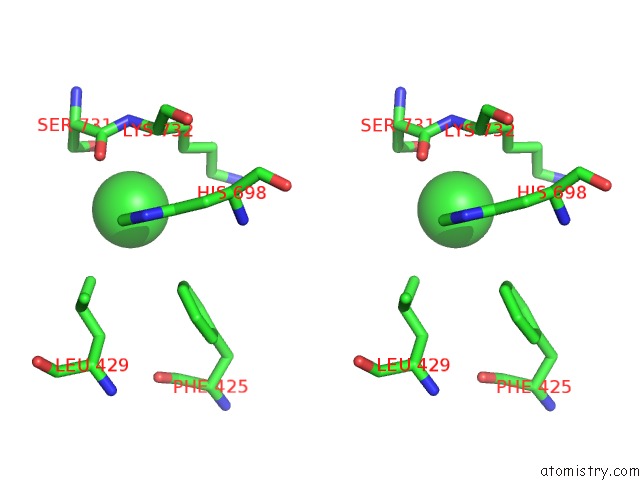
Chlorine binding site 4 out of 8 in 3ppc
Go back to
Chlorine binding site 4 out
of 8 in the Crystal Structure of the Candida Albicans Methionine Synthase By Surface Entropy Reduction, Tyrosine Variant with Zinc
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Crystal Structure of the Candida Albicans Methionine Synthase By Surface Entropy Reduction, Tyrosine Variant with Zinc within 5.0Å range:
|
Chlorine binding site 5 out of 8 in 3ppc
Go back to
Chlorine binding site 5 out
of 8 in the Crystal Structure of the Candida Albicans Methionine Synthase By Surface Entropy Reduction, Tyrosine Variant with Zinc
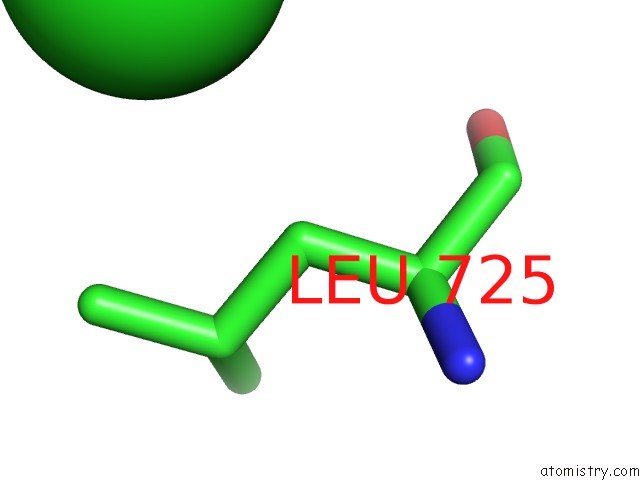
Mono view
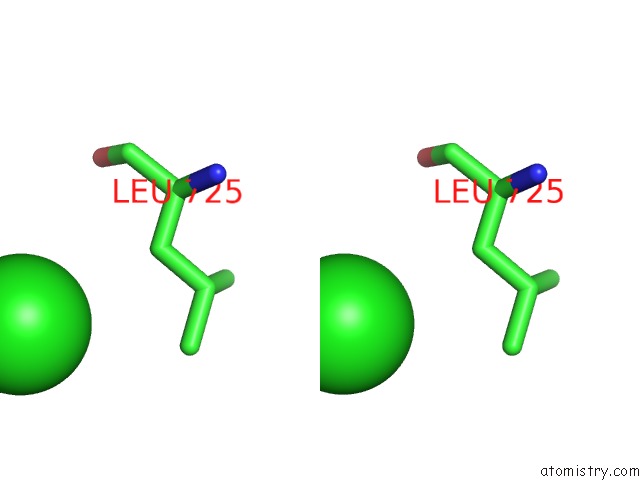
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Crystal Structure of the Candida Albicans Methionine Synthase By Surface Entropy Reduction, Tyrosine Variant with Zinc within 5.0Å range:
|
Chlorine binding site 6 out of 8 in 3ppc
Go back to
Chlorine binding site 6 out
of 8 in the Crystal Structure of the Candida Albicans Methionine Synthase By Surface Entropy Reduction, Tyrosine Variant with Zinc
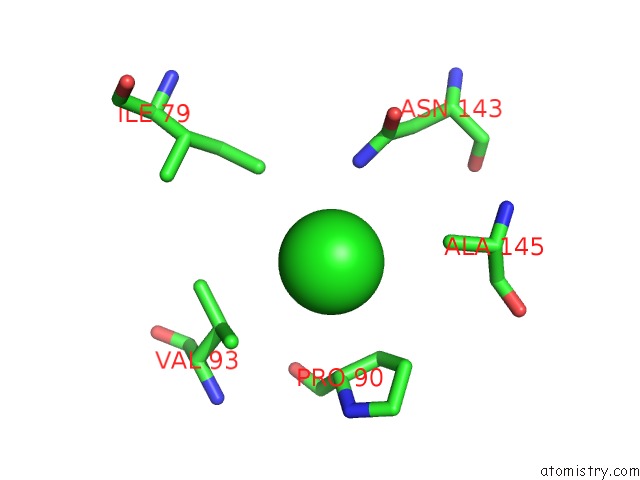
Mono view
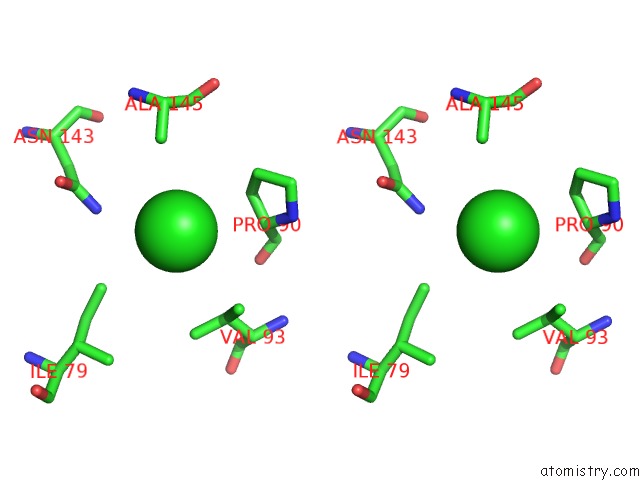
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of Crystal Structure of the Candida Albicans Methionine Synthase By Surface Entropy Reduction, Tyrosine Variant with Zinc within 5.0Å range:
|
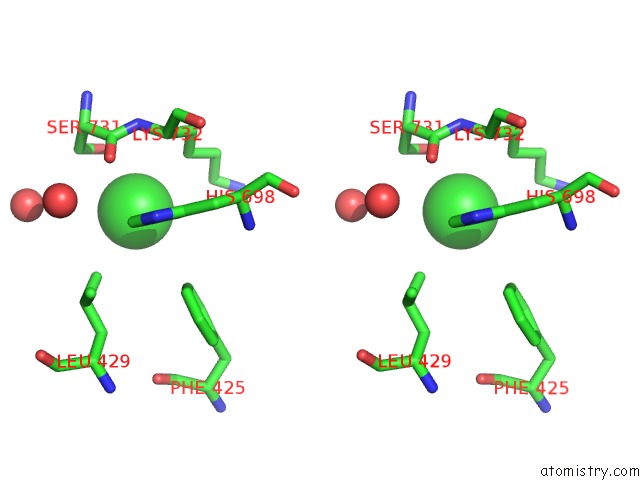
Chlorine binding site 7 out of 8 in 3ppc
Go back to
Chlorine binding site 7 out
of 8 in the Crystal Structure of the Candida Albicans Methionine Synthase By Surface Entropy Reduction, Tyrosine Variant with Zinc
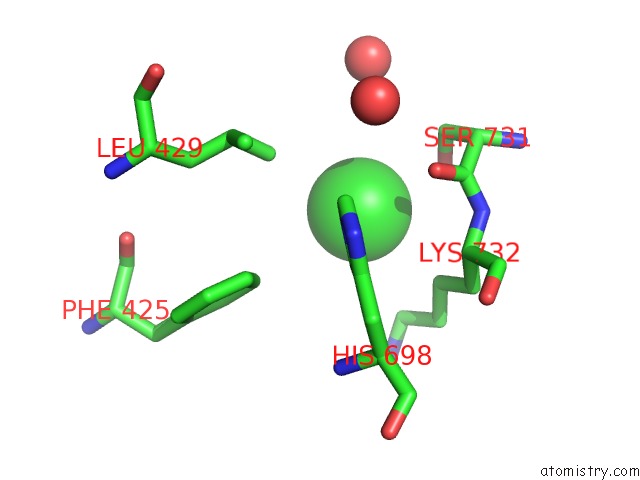
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 7 of Crystal Structure of the Candida Albicans Methionine Synthase By Surface Entropy Reduction, Tyrosine Variant with Zinc within 5.0Å range:
|
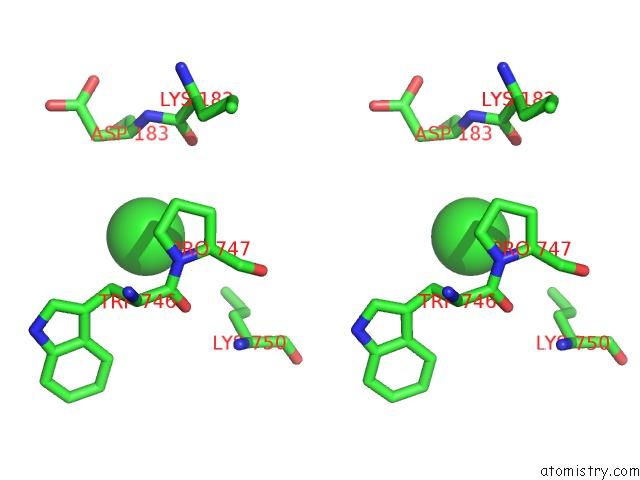
Chlorine binding site 8 out of 8 in 3ppc
Go back to
Chlorine binding site 8 out
of 8 in the Crystal Structure of the Candida Albicans Methionine Synthase By Surface Entropy Reduction, Tyrosine Variant with Zinc
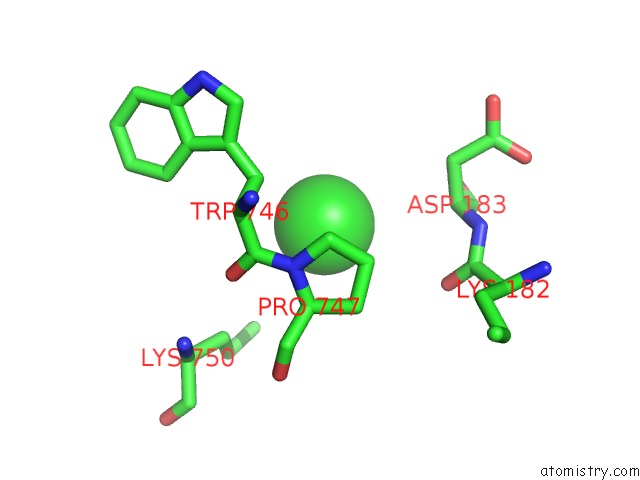
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 8 of Crystal Structure of the Candida Albicans Methionine Synthase By Surface Entropy Reduction, Tyrosine Variant with Zinc within 5.0Å range:
|
Reference:
D.Ubhi,
K.L.Kavanagh,
A.F.Monzingo,
J.D.Robertus.
Structure of Candida Albicans Methionine Synthase Determined By Employing Surface Residue Mutagenesis. Arch.Biochem.Biophys. V. 513 19 2011.
ISSN: ISSN 0003-9861
PubMed: 21689631
DOI: 10.1016/J.ABB.2011.06.002
Page generated: Sun Jul 21 02:33:19 2024
ISSN: ISSN 0003-9861
PubMed: 21689631
DOI: 10.1016/J.ABB.2011.06.002
Last articles
Cl in 2XIYCl in 2XHR
Cl in 2XIW
Cl in 2XIK
Cl in 2XEW
Cl in 2XGW
Cl in 2XFH
Cl in 2XGE
Cl in 2XGD
Cl in 2XFG