Chlorine »
PDB 4qgs-4qol »
4qnl »
Chlorine in PDB 4qnl: Crystal Structure of Tail Fiber Protein GP63.1 From E. Coli Phage G7C
Protein crystallography data
The structure of Crystal Structure of Tail Fiber Protein GP63.1 From E. Coli Phage G7C, PDB code: 4qnl
was solved by
C.Riccio,
C.Browning,
N.Prokhorov,
A.Letarov,
P.G.Leiman,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 64.96 / 2.41 |
| Space group | H 3 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 92.773, 92.773, 551.926, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 15.9 / 22.1 |
Other elements in 4qnl:
The structure of Crystal Structure of Tail Fiber Protein GP63.1 From E. Coli Phage G7C also contains other interesting chemical elements:
| Zinc | (Zn) | 1 atom |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of Tail Fiber Protein GP63.1 From E. Coli Phage G7C
(pdb code 4qnl). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 4 binding sites of Chlorine where determined in the Crystal Structure of Tail Fiber Protein GP63.1 From E. Coli Phage G7C, PDB code: 4qnl:
Jump to Chlorine binding site number: 1; 2; 3; 4;
In total 4 binding sites of Chlorine where determined in the Crystal Structure of Tail Fiber Protein GP63.1 From E. Coli Phage G7C, PDB code: 4qnl:
Jump to Chlorine binding site number: 1; 2; 3; 4;
Chlorine binding site 1 out of 4 in 4qnl
Go back to
Chlorine binding site 1 out
of 4 in the Crystal Structure of Tail Fiber Protein GP63.1 From E. Coli Phage G7C
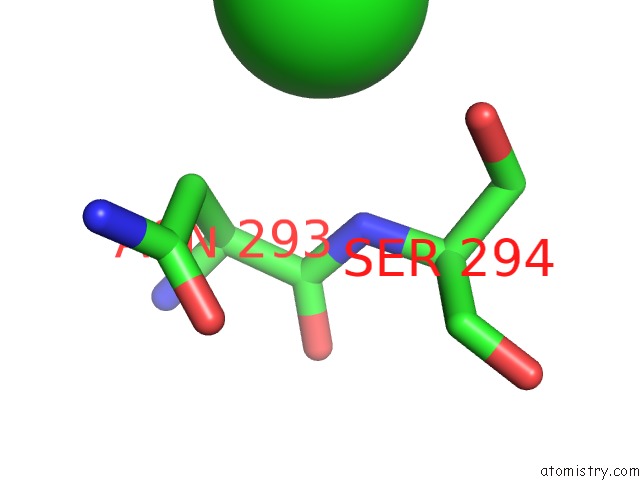
Mono view
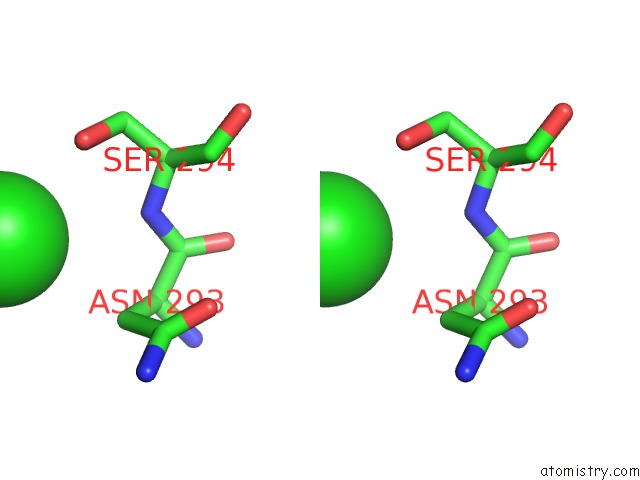
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Tail Fiber Protein GP63.1 From E. Coli Phage G7C within 5.0Å range:
|
Chlorine binding site 2 out of 4 in 4qnl
Go back to
Chlorine binding site 2 out
of 4 in the Crystal Structure of Tail Fiber Protein GP63.1 From E. Coli Phage G7C
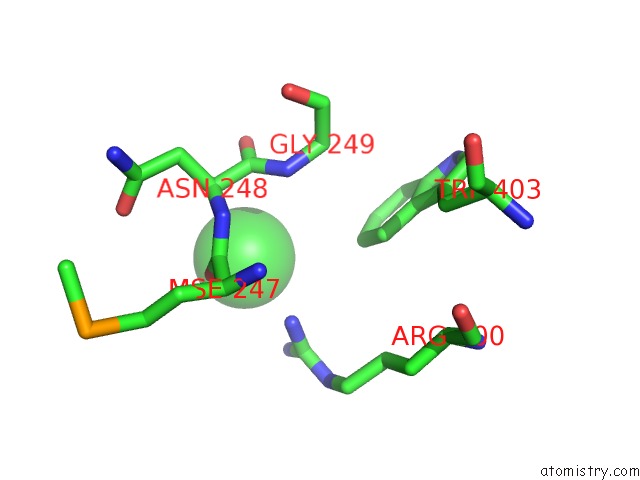
Mono view
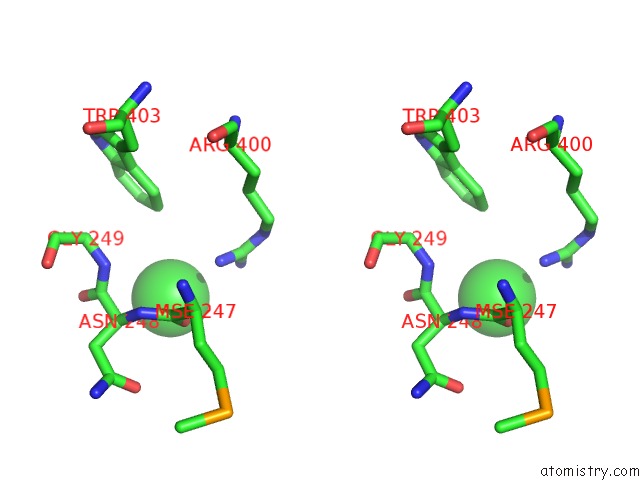
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of Tail Fiber Protein GP63.1 From E. Coli Phage G7C within 5.0Å range:
|
Chlorine binding site 3 out of 4 in 4qnl
Go back to
Chlorine binding site 3 out
of 4 in the Crystal Structure of Tail Fiber Protein GP63.1 From E. Coli Phage G7C
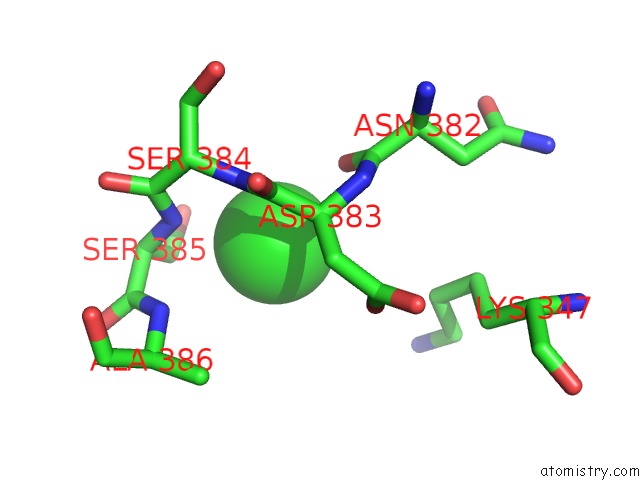
Mono view
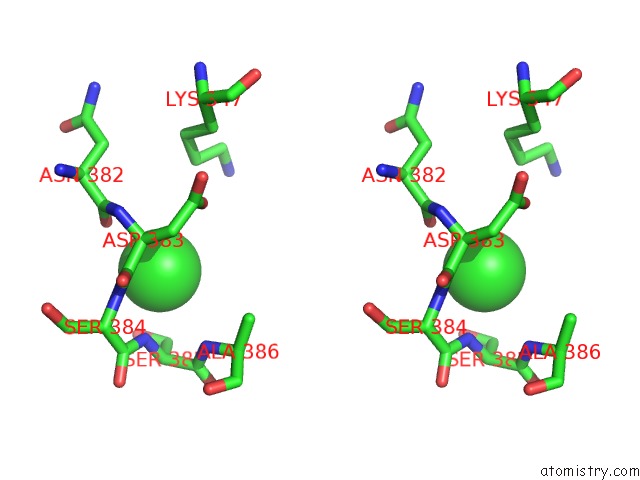
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Crystal Structure of Tail Fiber Protein GP63.1 From E. Coli Phage G7C within 5.0Å range:
|
Chlorine binding site 4 out of 4 in 4qnl
Go back to
Chlorine binding site 4 out
of 4 in the Crystal Structure of Tail Fiber Protein GP63.1 From E. Coli Phage G7C
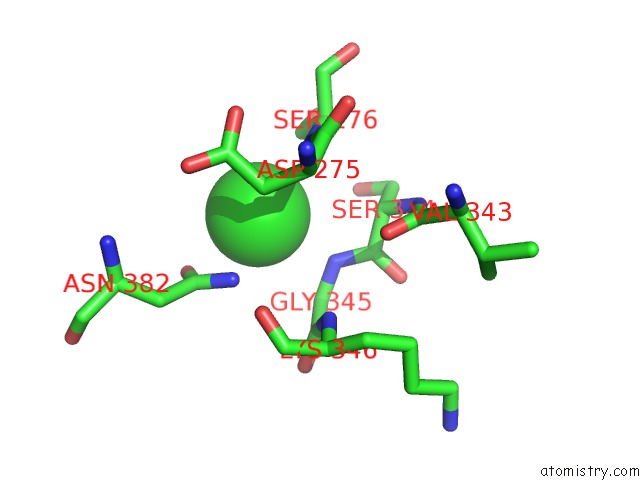
Mono view
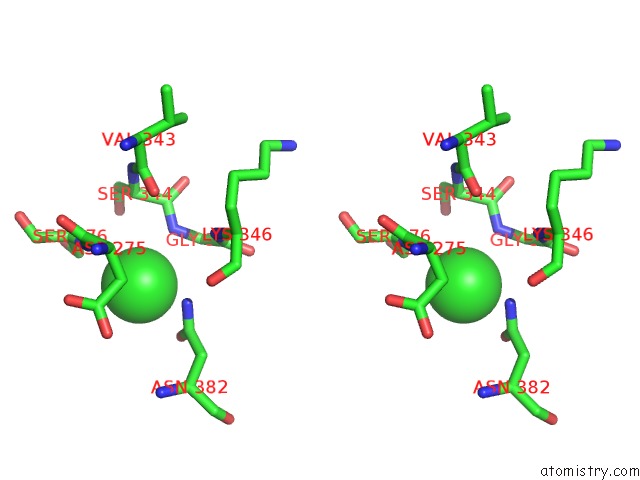
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Crystal Structure of Tail Fiber Protein GP63.1 From E. Coli Phage G7C within 5.0Å range:
|
Reference:
N.S.Prokhorov,
C.Riccio,
E.L.Zdorovenko,
M.M.Shneider,
C.Browning,
Y.A.Knirel,
P.G.Leiman,
A.V.Letarov.
Function of Bacteriophage G7C Esterase Tailspike in Host Cell Adsorption. Mol. Microbiol. V. 105 385 2017.
ISSN: ESSN 1365-2958
PubMed: 28513100
DOI: 10.1111/MMI.13710
Page generated: Fri Jul 26 00:34:46 2024
ISSN: ESSN 1365-2958
PubMed: 28513100
DOI: 10.1111/MMI.13710
Last articles
Cl in 2XNECl in 2XN5
Cl in 2XMS
Cl in 2XMG
Cl in 2XMD
Cl in 2XML
Cl in 2XMM
Cl in 2XMK
Cl in 2XMJ
Cl in 2XMB