Chlorine »
PDB 5eaf-5eic »
5eeh »
Chlorine in PDB 5eeh: Crystal Structure of Carminomycin-4-O-Methyltransferase Dnrk in Complex with Sah and 2-Chloro-4-Nitrophenol
Enzymatic activity of Crystal Structure of Carminomycin-4-O-Methyltransferase Dnrk in Complex with Sah and 2-Chloro-4-Nitrophenol
All present enzymatic activity of Crystal Structure of Carminomycin-4-O-Methyltransferase Dnrk in Complex with Sah and 2-Chloro-4-Nitrophenol:
2.1.1.292;
2.1.1.292;
Protein crystallography data
The structure of Crystal Structure of Carminomycin-4-O-Methyltransferase Dnrk in Complex with Sah and 2-Chloro-4-Nitrophenol, PDB code: 5eeh
was solved by
F.Wang,
S.Singh,
J.S.Thorson,
G.N.Phillips Jr.,
Enzyme Discovery Fornatural Product Biosynthesis (Natpro),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.50 / 1.82 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 123.347, 110.678, 116.869, 90.00, 120.24, 90.00 |
| R / Rfree (%) | 15.4 / 18.6 |
Chlorine Binding Sites:
Pages:
>>> Page 1 <<< Page 2, Binding sites: 11 - 15;Binding sites:
The binding sites of Chlorine atom in the Crystal Structure of Carminomycin-4-O-Methyltransferase Dnrk in Complex with Sah and 2-Chloro-4-Nitrophenol (pdb code 5eeh). This binding sites where shown within 5.0 Angstroms radius around Chlorine atom.In total 15 binding sites of Chlorine where determined in the Crystal Structure of Carminomycin-4-O-Methyltransferase Dnrk in Complex with Sah and 2-Chloro-4-Nitrophenol, PDB code: 5eeh:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
Chlorine binding site 1 out of 15 in 5eeh
Go back to
Chlorine binding site 1 out
of 15 in the Crystal Structure of Carminomycin-4-O-Methyltransferase Dnrk in Complex with Sah and 2-Chloro-4-Nitrophenol
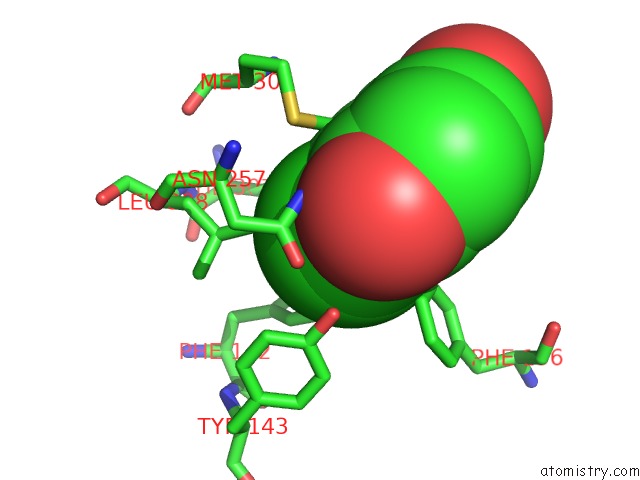
Mono view
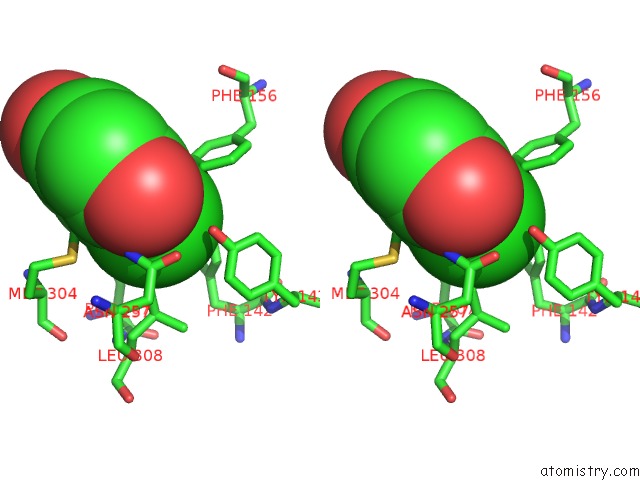
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Carminomycin-4-O-Methyltransferase Dnrk in Complex with Sah and 2-Chloro-4-Nitrophenol within 5.0Å range:
|
Chlorine binding site 2 out of 15 in 5eeh
Go back to
Chlorine binding site 2 out
of 15 in the Crystal Structure of Carminomycin-4-O-Methyltransferase Dnrk in Complex with Sah and 2-Chloro-4-Nitrophenol
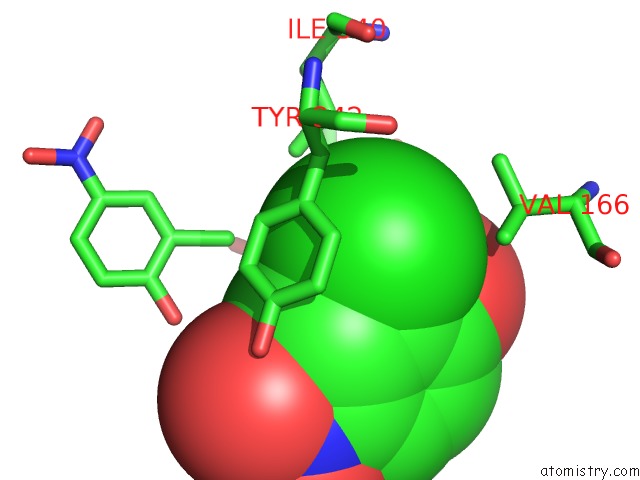
Mono view
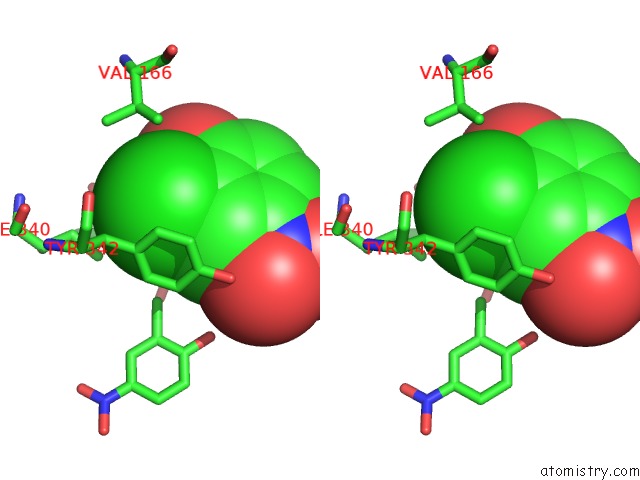
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of Carminomycin-4-O-Methyltransferase Dnrk in Complex with Sah and 2-Chloro-4-Nitrophenol within 5.0Å range:
|
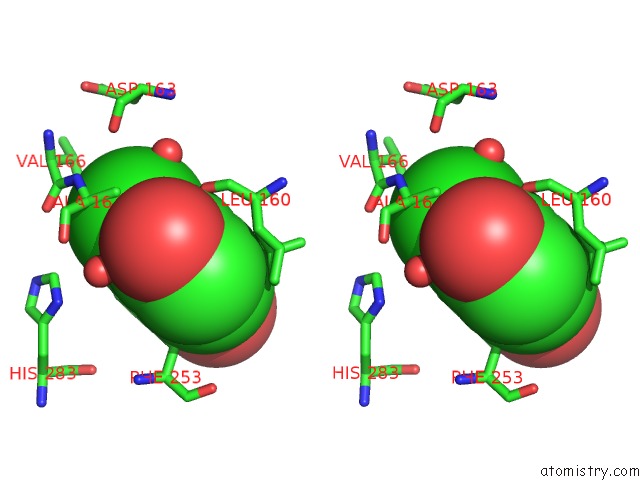
Chlorine binding site 3 out of 15 in 5eeh
Go back to
Chlorine binding site 3 out
of 15 in the Crystal Structure of Carminomycin-4-O-Methyltransferase Dnrk in Complex with Sah and 2-Chloro-4-Nitrophenol
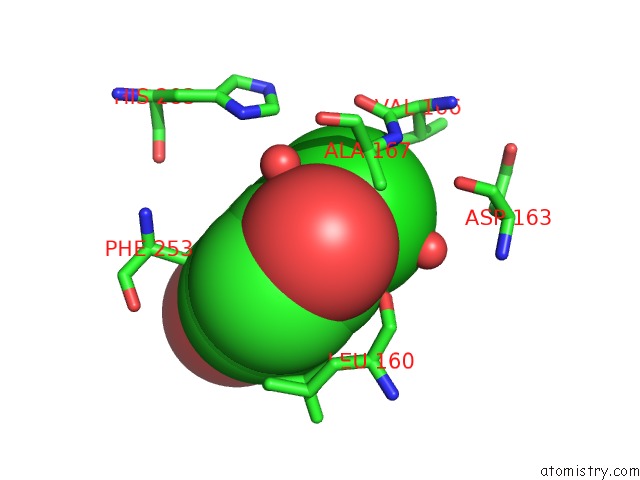
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Crystal Structure of Carminomycin-4-O-Methyltransferase Dnrk in Complex with Sah and 2-Chloro-4-Nitrophenol within 5.0Å range:
|
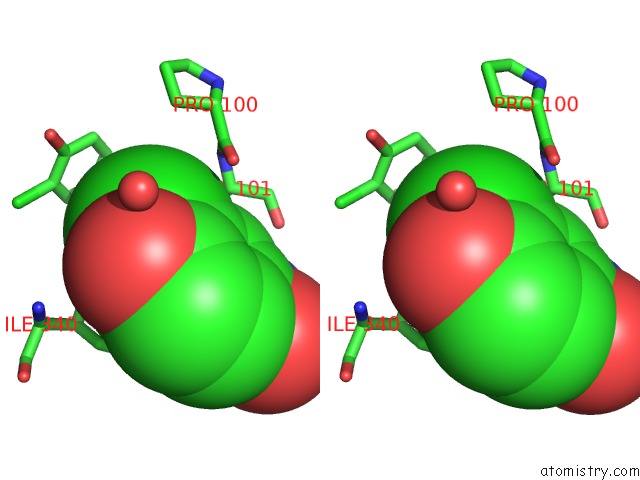
Chlorine binding site 4 out of 15 in 5eeh
Go back to
Chlorine binding site 4 out
of 15 in the Crystal Structure of Carminomycin-4-O-Methyltransferase Dnrk in Complex with Sah and 2-Chloro-4-Nitrophenol
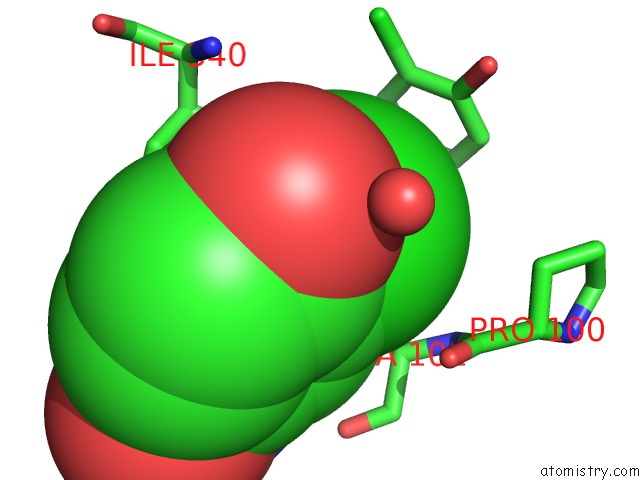
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Crystal Structure of Carminomycin-4-O-Methyltransferase Dnrk in Complex with Sah and 2-Chloro-4-Nitrophenol within 5.0Å range:
|
Chlorine binding site 5 out of 15 in 5eeh
Go back to
Chlorine binding site 5 out
of 15 in the Crystal Structure of Carminomycin-4-O-Methyltransferase Dnrk in Complex with Sah and 2-Chloro-4-Nitrophenol
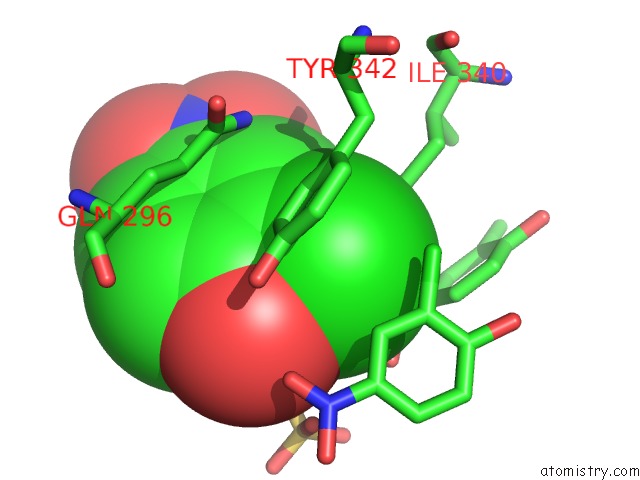
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Crystal Structure of Carminomycin-4-O-Methyltransferase Dnrk in Complex with Sah and 2-Chloro-4-Nitrophenol within 5.0Å range:
|
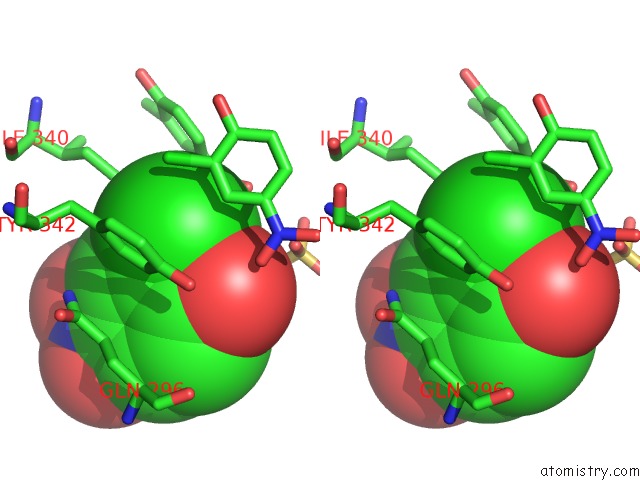
Chlorine binding site 6 out of 15 in 5eeh
Go back to
Chlorine binding site 6 out
of 15 in the Crystal Structure of Carminomycin-4-O-Methyltransferase Dnrk in Complex with Sah and 2-Chloro-4-Nitrophenol
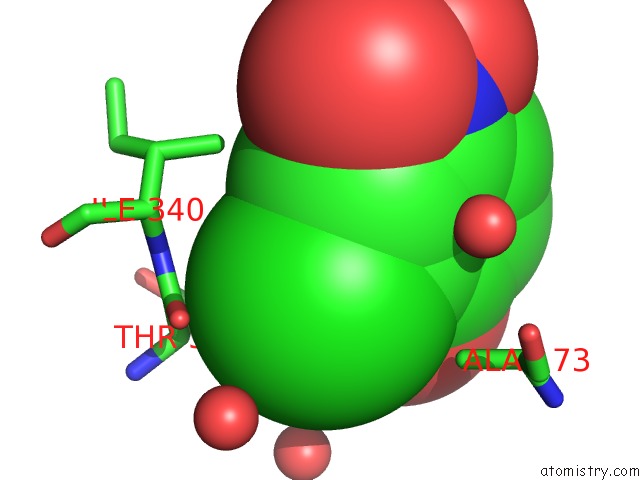
Mono view
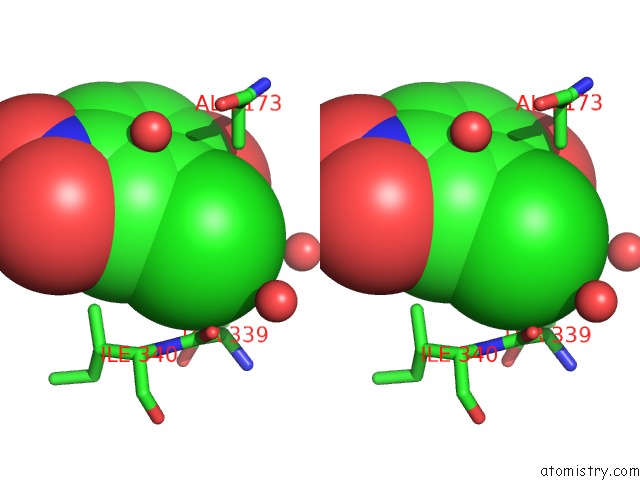
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of Crystal Structure of Carminomycin-4-O-Methyltransferase Dnrk in Complex with Sah and 2-Chloro-4-Nitrophenol within 5.0Å range:
|
Chlorine binding site 7 out of 15 in 5eeh
Go back to
Chlorine binding site 7 out
of 15 in the Crystal Structure of Carminomycin-4-O-Methyltransferase Dnrk in Complex with Sah and 2-Chloro-4-Nitrophenol
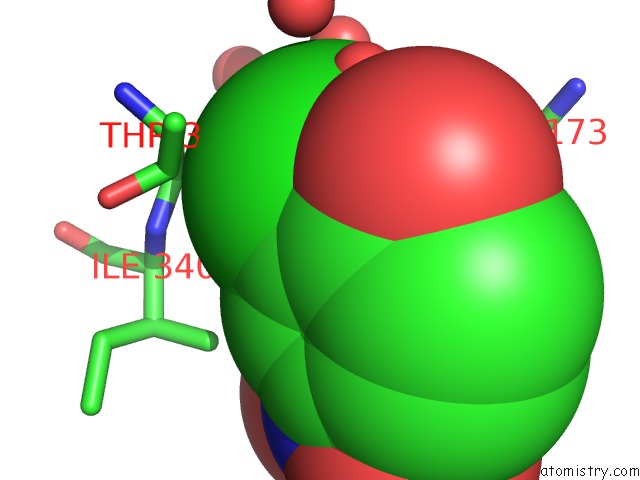
Mono view
Stereo pair view

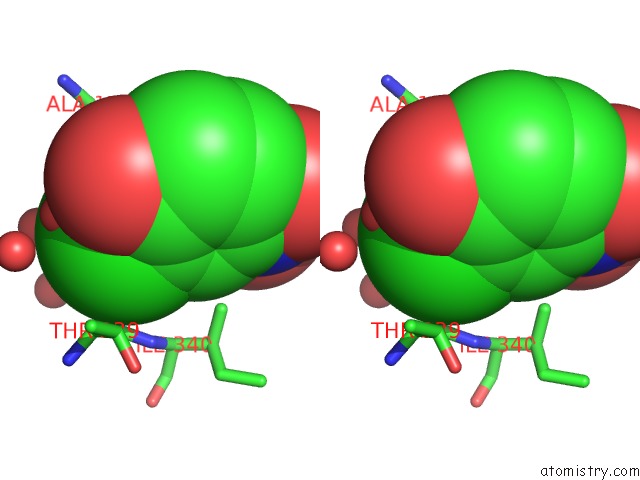
Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 7 of Crystal Structure of Carminomycin-4-O-Methyltransferase Dnrk in Complex with Sah and 2-Chloro-4-Nitrophenol within 5.0Å range:
|
Chlorine binding site 8 out of 15 in 5eeh
Go back to
Chlorine binding site 8 out
of 15 in the Crystal Structure of Carminomycin-4-O-Methyltransferase Dnrk in Complex with Sah and 2-Chloro-4-Nitrophenol
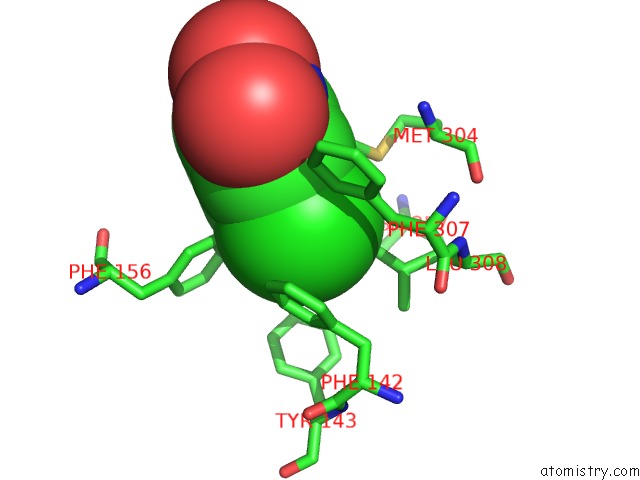
Mono view
Stereo pair view

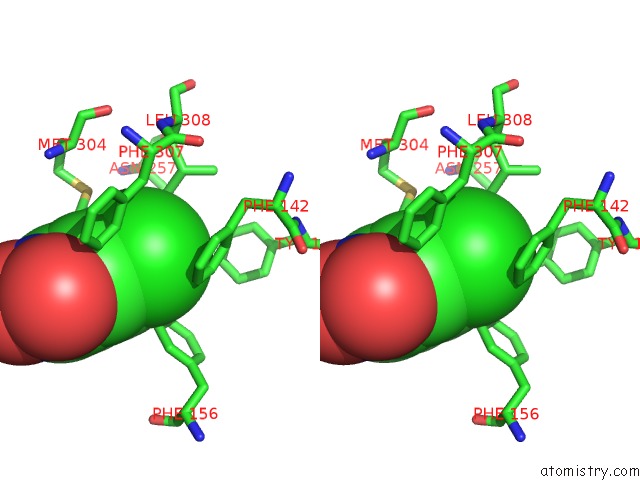
Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 8 of Crystal Structure of Carminomycin-4-O-Methyltransferase Dnrk in Complex with Sah and 2-Chloro-4-Nitrophenol within 5.0Å range:
|
Chlorine binding site 9 out of 15 in 5eeh
Go back to
Chlorine binding site 9 out
of 15 in the Crystal Structure of Carminomycin-4-O-Methyltransferase Dnrk in Complex with Sah and 2-Chloro-4-Nitrophenol
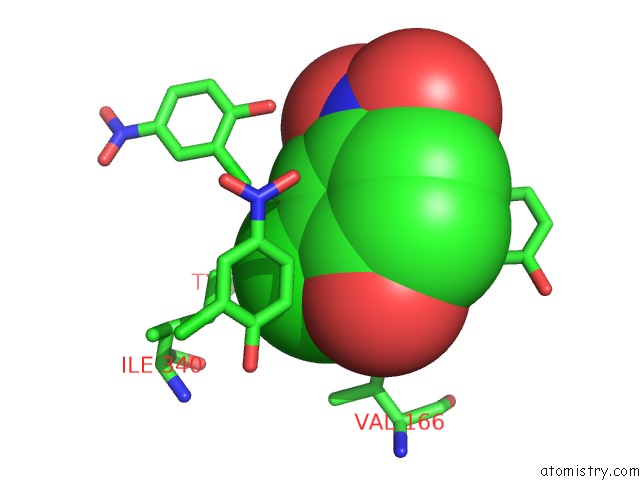
Mono view
Stereo pair view

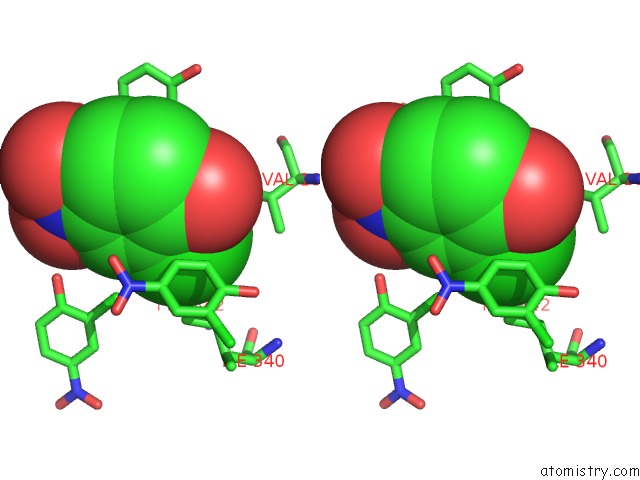
Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 9 of Crystal Structure of Carminomycin-4-O-Methyltransferase Dnrk in Complex with Sah and 2-Chloro-4-Nitrophenol within 5.0Å range:
|
Chlorine binding site 10 out of 15 in 5eeh
Go back to
Chlorine binding site 10 out
of 15 in the Crystal Structure of Carminomycin-4-O-Methyltransferase Dnrk in Complex with Sah and 2-Chloro-4-Nitrophenol
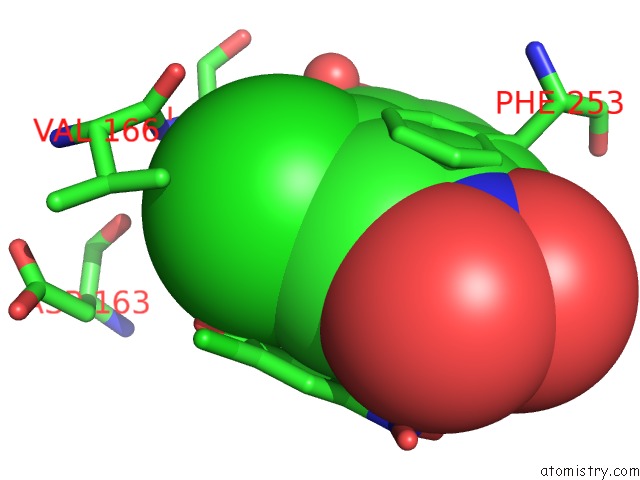
Mono view
Stereo pair view

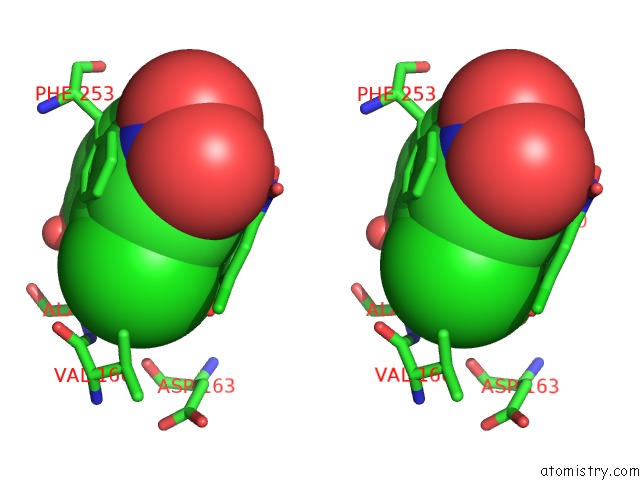
Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 10 of Crystal Structure of Carminomycin-4-O-Methyltransferase Dnrk in Complex with Sah and 2-Chloro-4-Nitrophenol within 5.0Å range:
|
Reference:
T.D.Huber,
F.Wang,
S.Singh,
B.R.Johnson,
J.Zhang,
M.Sunkara,
S.G.Van Lanen,
A.J.Morris,
G.N.Phillips,
J.S.Thorson.
Functional Adomet Isosteres Resistant to Classical Adomet Degradation Pathways. Acs Chem.Biol. V. 11 2484 2016.
ISSN: ESSN 1554-8937
PubMed: 27351335
DOI: 10.1021/ACSCHEMBIO.6B00348
Page generated: Sat Jul 12 01:40:03 2025
ISSN: ESSN 1554-8937
PubMed: 27351335
DOI: 10.1021/ACSCHEMBIO.6B00348
Last articles
Cl in 7ZD1Cl in 7ZD0
Cl in 7ZBV
Cl in 7ZCB
Cl in 7ZCY
Cl in 7ZBD
Cl in 7ZBB
Cl in 7ZBA
Cl in 7Z53
Cl in 7ZB9