Chlorine »
PDB 5kw1-5l5a »
5kxm »
Chlorine in PDB 5kxm: Hen Egg White Lysozyme at 100K, Data Set 3
Enzymatic activity of Hen Egg White Lysozyme at 100K, Data Set 3
All present enzymatic activity of Hen Egg White Lysozyme at 100K, Data Set 3:
3.2.1.17;
3.2.1.17;
Protein crystallography data
The structure of Hen Egg White Lysozyme at 100K, Data Set 3, PDB code: 5kxm
was solved by
S.Russi,
A.Gonzalez,
L.R.Kenner,
D.A.Keedy,
J.S.Fraser,
H.Van Den Bedem,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 37.20 / 1.20 |
| Space group | P 43 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 77.289, 77.289, 37.205, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 15.2 / 18 |
Other elements in 5kxm:
The structure of Hen Egg White Lysozyme at 100K, Data Set 3 also contains other interesting chemical elements:
| Sodium | (Na) | 2 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Hen Egg White Lysozyme at 100K, Data Set 3
(pdb code 5kxm). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 3 binding sites of Chlorine where determined in the Hen Egg White Lysozyme at 100K, Data Set 3, PDB code: 5kxm:
Jump to Chlorine binding site number: 1; 2; 3;
In total 3 binding sites of Chlorine where determined in the Hen Egg White Lysozyme at 100K, Data Set 3, PDB code: 5kxm:
Jump to Chlorine binding site number: 1; 2; 3;
Chlorine binding site 1 out of 3 in 5kxm
Go back to
Chlorine binding site 1 out
of 3 in the Hen Egg White Lysozyme at 100K, Data Set 3
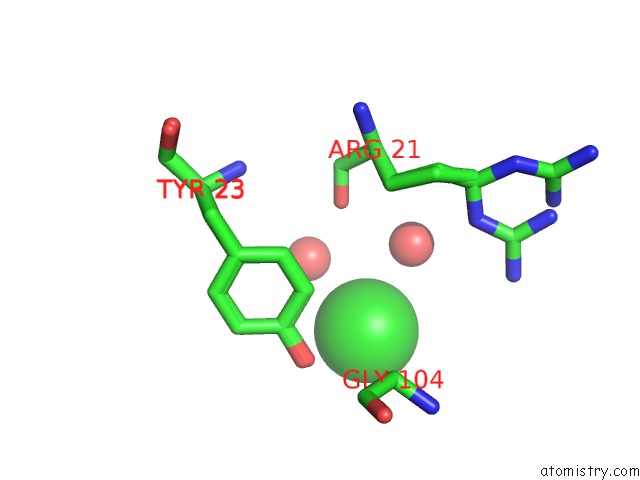
Mono view
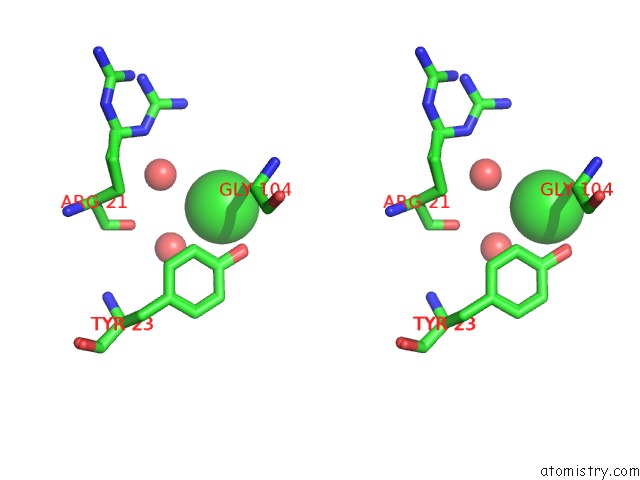
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Hen Egg White Lysozyme at 100K, Data Set 3 within 5.0Å range:
|
Chlorine binding site 2 out of 3 in 5kxm
Go back to
Chlorine binding site 2 out
of 3 in the Hen Egg White Lysozyme at 100K, Data Set 3
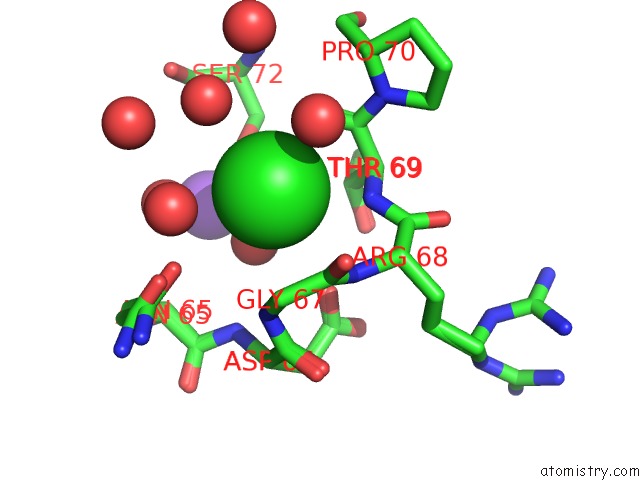
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Hen Egg White Lysozyme at 100K, Data Set 3 within 5.0Å range:
|
Chlorine binding site 3 out of 3 in 5kxm
Go back to
Chlorine binding site 3 out
of 3 in the Hen Egg White Lysozyme at 100K, Data Set 3
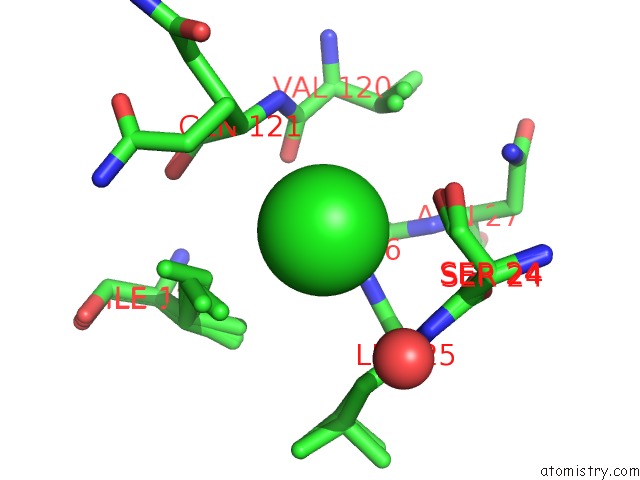
Mono view
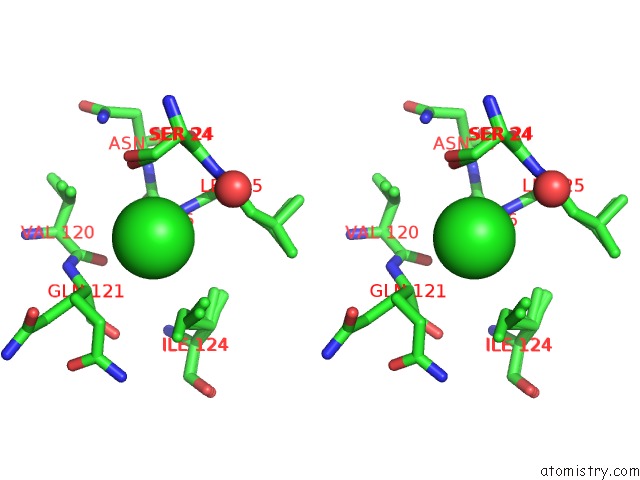
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Hen Egg White Lysozyme at 100K, Data Set 3 within 5.0Å range:
|
Reference:
S.Russi,
A.Gonzalez,
L.R.Kenner,
D.A.Keedy,
J.S.Fraser,
H.Van Den Bedem.
Conformational Variation of Proteins at Room Temperature Is Not Dominated By Radiation Damage. J Synchrotron Radiat V. 24 73 2017.
ISSN: ESSN 1600-5775
PubMed: 28009548
DOI: 10.1107/S1600577516017343
Page generated: Sat Jul 12 04:15:31 2025
ISSN: ESSN 1600-5775
PubMed: 28009548
DOI: 10.1107/S1600577516017343
Last articles
Cl in 7LV7Cl in 7LV5
Cl in 7LUN
Cl in 7LUF
Cl in 7LTJ
Cl in 7LTB
Cl in 7LTP
Cl in 7LT4
Cl in 7LRQ
Cl in 7LRO