Chlorine »
PDB 6vx6-6w7p »
6w2x »
Chlorine in PDB 6w2x: Cryoem Structure of Inactive Gabab Heterodimer
Other elements in 6w2x:
The structure of Cryoem Structure of Inactive Gabab Heterodimer also contains other interesting chemical elements:
| Magnesium | (Mg) | 1 atom |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Cryoem Structure of Inactive Gabab Heterodimer
(pdb code 6w2x). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Cryoem Structure of Inactive Gabab Heterodimer, PDB code: 6w2x:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Cryoem Structure of Inactive Gabab Heterodimer, PDB code: 6w2x:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 6w2x
Go back to
Chlorine binding site 1 out
of 2 in the Cryoem Structure of Inactive Gabab Heterodimer
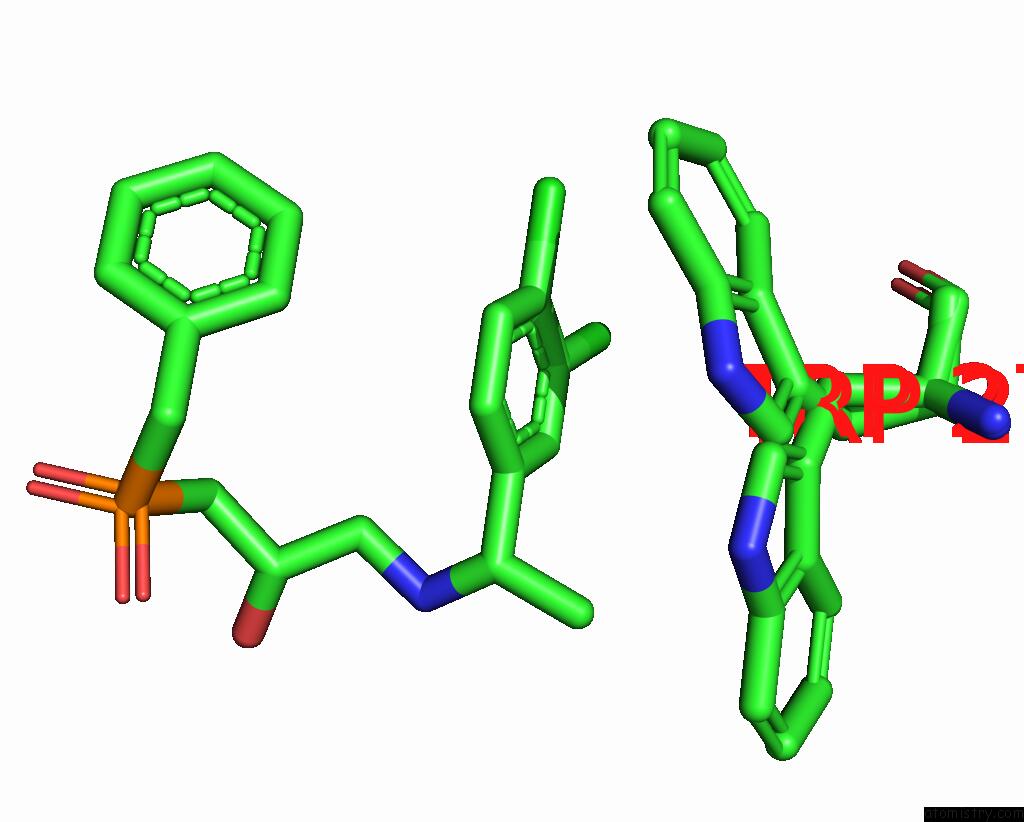
Mono view
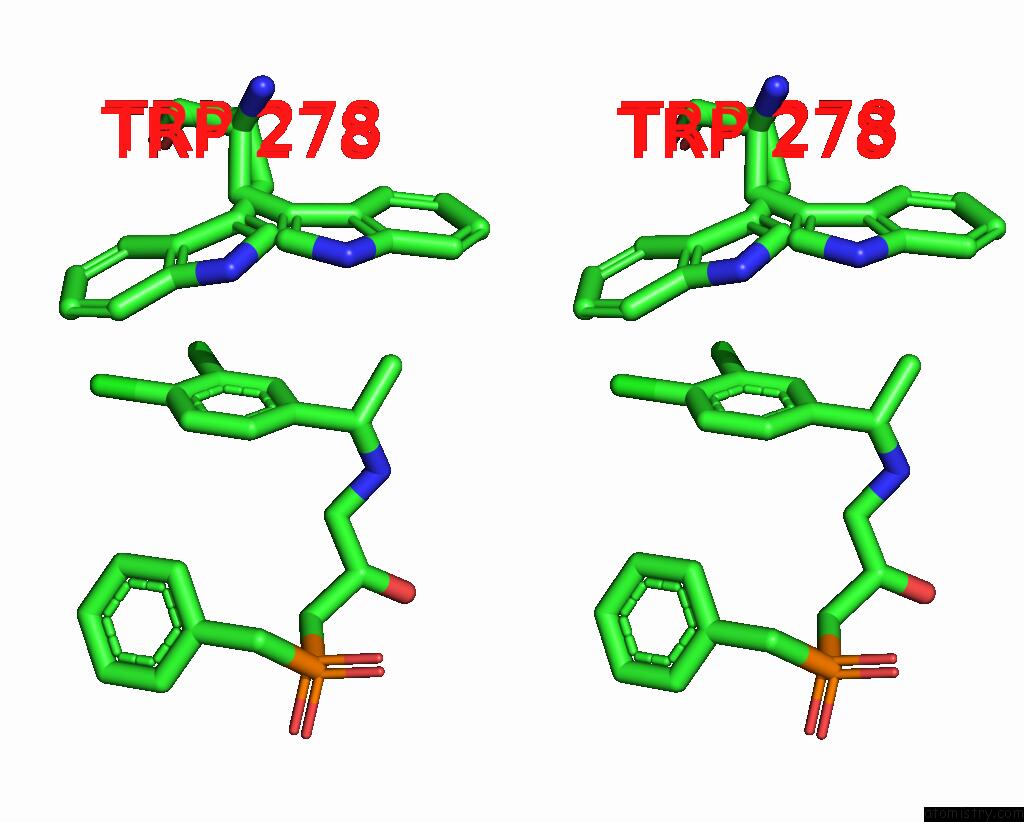
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Cryoem Structure of Inactive Gabab Heterodimer within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 6w2x
Go back to
Chlorine binding site 2 out
of 2 in the Cryoem Structure of Inactive Gabab Heterodimer
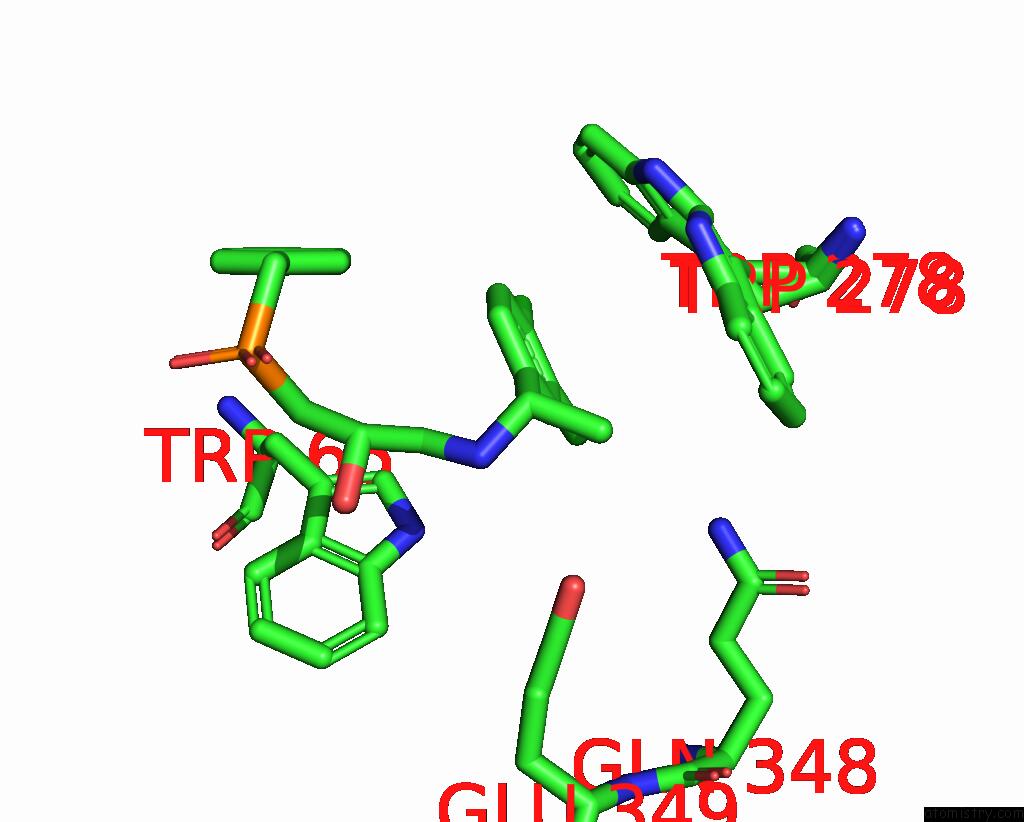
Mono view
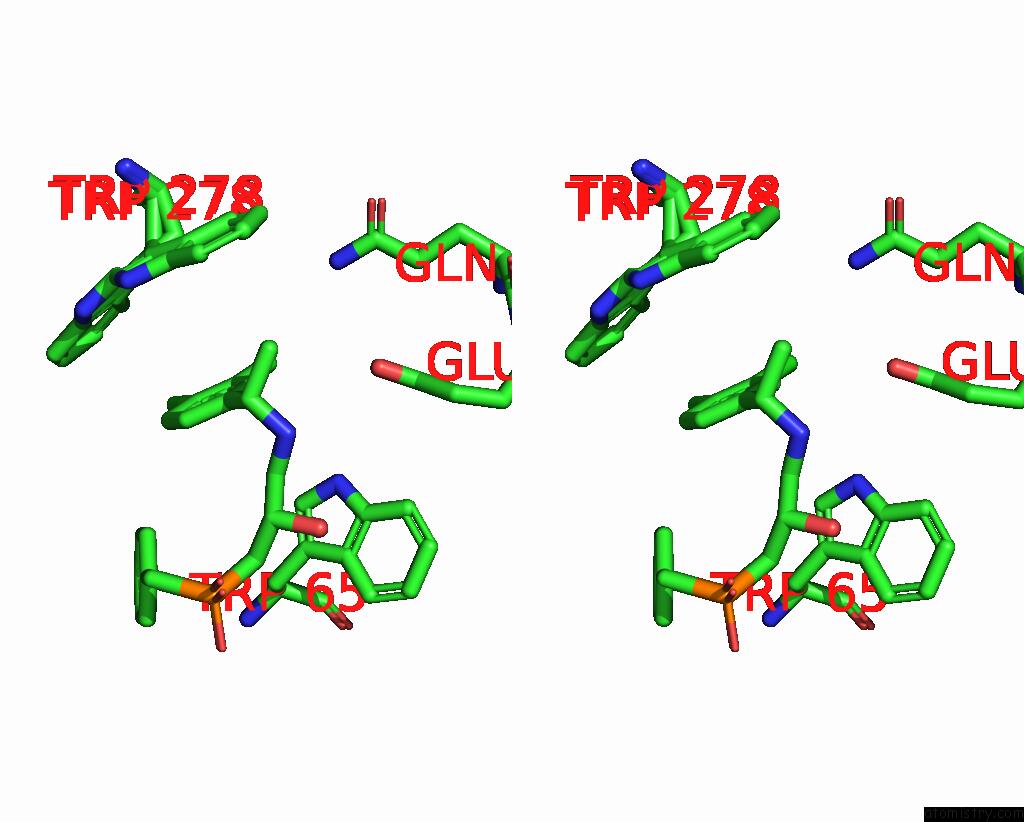
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Cryoem Structure of Inactive Gabab Heterodimer within 5.0Å range:
|
Reference:
M.M.Papasergi-Scott,
M.J.Robertson,
A.B.Seven,
O.Panova,
J.M.Mathiesen,
G.Skiniotis.
Structures of Metabotropic Gababreceptor. Nature V. 584 310 2020.
ISSN: ESSN 1476-4687
PubMed: 32580208
DOI: 10.1038/S41586-020-2469-4
Page generated: Sat Jul 12 21:07:13 2025
ISSN: ESSN 1476-4687
PubMed: 32580208
DOI: 10.1038/S41586-020-2469-4
Last articles
Cl in 7ZBACl in 7Z53
Cl in 7ZB9
Cl in 7Z93
Cl in 7Z88
Cl in 7Z8Y
Cl in 7Z8W
Cl in 7Z87
Cl in 7Z7B
Cl in 7Z5N