Chlorine »
PDB 7vkh-7vzz »
7vug »
Chlorine in PDB 7vug: Cryo-Em Structure of A Class A Orphan Gpcr in Complex with Gi
Enzymatic activity of Cryo-Em Structure of A Class A Orphan Gpcr in Complex with Gi
All present enzymatic activity of Cryo-Em Structure of A Class A Orphan Gpcr in Complex with Gi:
3.2.1.8;
3.2.1.8;
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Cryo-Em Structure of A Class A Orphan Gpcr in Complex with Gi
(pdb code 7vug). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Cryo-Em Structure of A Class A Orphan Gpcr in Complex with Gi, PDB code: 7vug:
In total only one binding site of Chlorine was determined in the Cryo-Em Structure of A Class A Orphan Gpcr in Complex with Gi, PDB code: 7vug:
Chlorine binding site 1 out of 1 in 7vug
Go back to
Chlorine binding site 1 out
of 1 in the Cryo-Em Structure of A Class A Orphan Gpcr in Complex with Gi
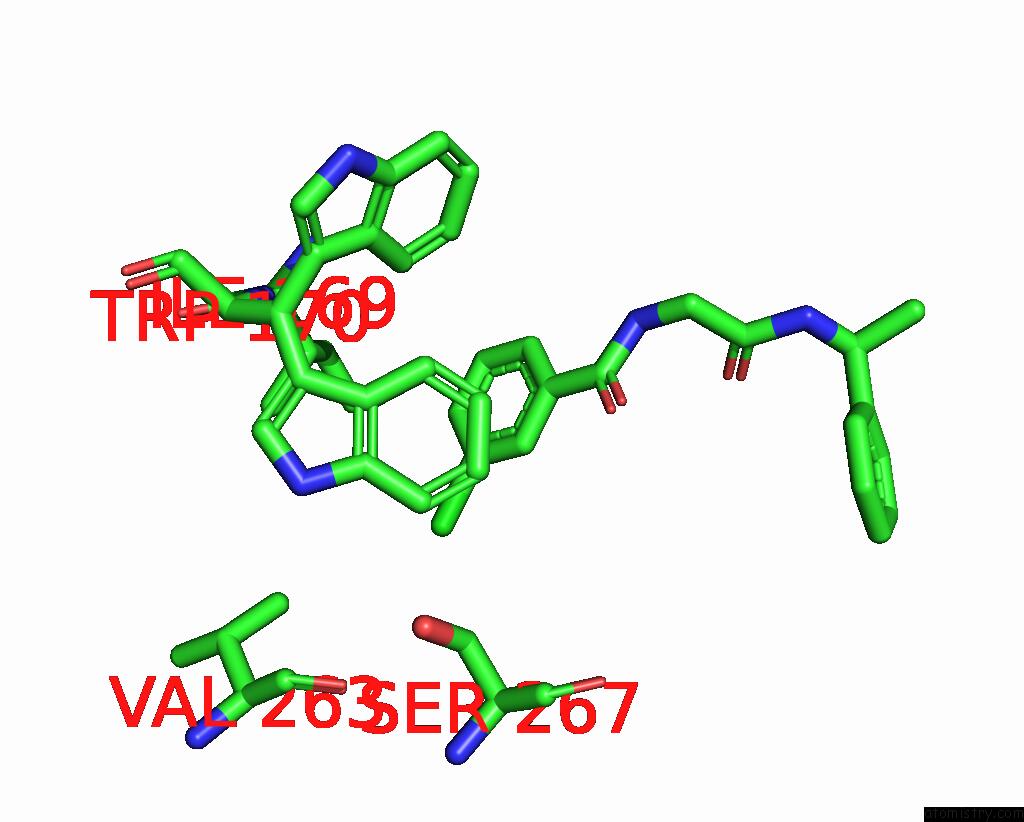
Mono view
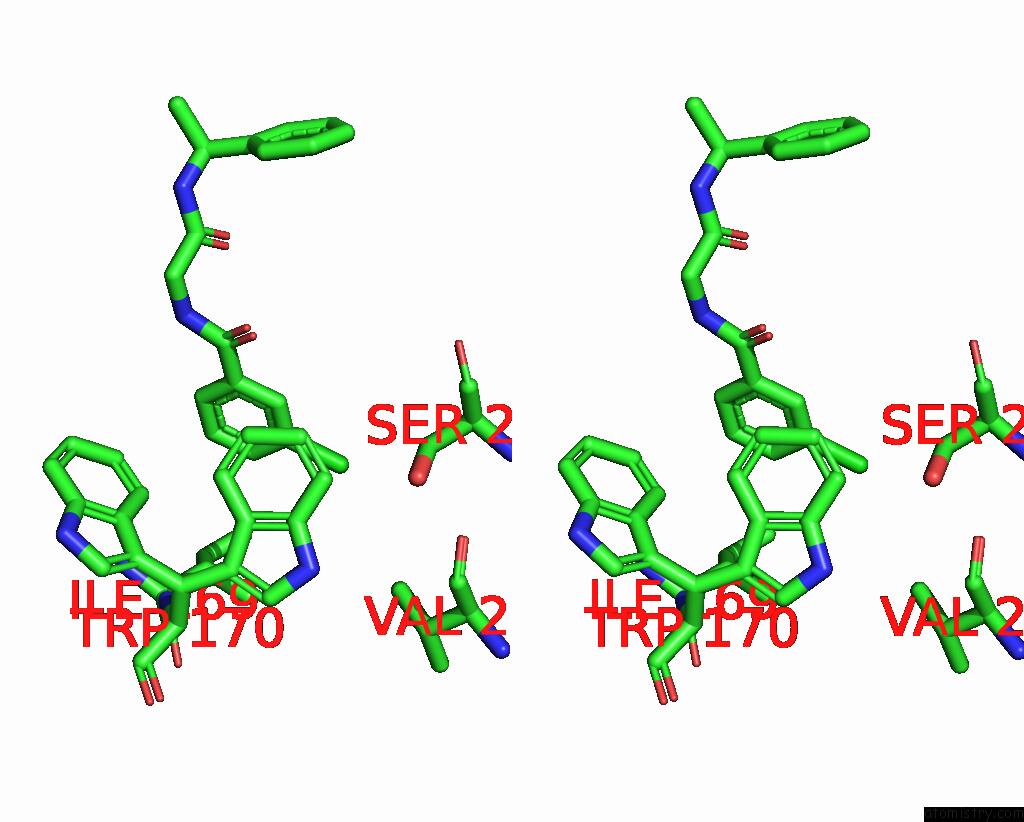
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Cryo-Em Structure of A Class A Orphan Gpcr in Complex with Gi within 5.0Å range:
|
Reference:
Y.Zhou,
H.Daver,
B.Trapkov,
L.Wu,
M.Wu,
K.Harpsoe,
P.R.Gentry,
K.Liu,
M.Larionova,
J.Liu,
N.Chen,
H.Brauner-Osborne,
D.E.Gloriam,
T.Hua,
Z.J.Liu.
Molecular Insights Into Ligand Recognition and G Protein Coupling of the Neuromodulatory Orphan Receptor GPR139. Cell Res. 2021.
ISSN: ISSN 1001-0602
PubMed: 34916631
DOI: 10.1038/S41422-021-00591-W
Page generated: Sun Jul 13 08:11:10 2025
ISSN: ISSN 1001-0602
PubMed: 34916631
DOI: 10.1038/S41422-021-00591-W
Last articles
Cl in 7ZNPCl in 7ZJV
Cl in 7ZLT
Cl in 7ZKS
Cl in 7ZKX
Cl in 7ZKR
Cl in 7ZJU
Cl in 7ZIV
Cl in 7ZJT
Cl in 7ZIK